Time Calibrated Phylogenies
Construction and evaluation of time calibrated trees for each serotype
Serotype A
Check tree model statistics the get_tracer_stats() function calculates similar summary statistics as the Tracer software typically used with BEAST.
Hide code
check_stats <- get_tracer_stats(here("local/beast/a_1/sero_a.log.txt"))
keep_stats <- c("joint", "prior", "likelihood", "treeModel.rootHeight", "age.root.",
"treeLength", "tmrca.fmd_a_rev.", "clock.rate")
check_stats %>%
filter(Parameter %in% keep_stats) %>%
gt() %>%
tab_header(
title = md("Serotype A Stats")) %>%
cols_width(starts_with("Parameter") ~ px(200),
starts_with("label") ~ px(60),
everything() ~ px(95)) %>%
tab_options(table.font.size = "small",
row_group.font.size = "small",
stub.font.size = "small",
column_labels.font.size = "medium",
heading.title.font.size = "large",
data_row.padding = px(2),
heading.title.font.weight = "bold",
column_labels.font.weight = "bold") %>%
opt_stylize(style = 6, color = 'gray')| Serotype A Stats | |||||
| Parameter | Mean | Median | Q_0.025 | Q_0.975 | ESS |
|---|---|---|---|---|---|
Load MCC tree
Hide code
sero_A.tree <- read.nexus(here("local/beast/a_1/sero_a.mcc.tre"))
plot_time_tree(sero_A.tree, check_stats)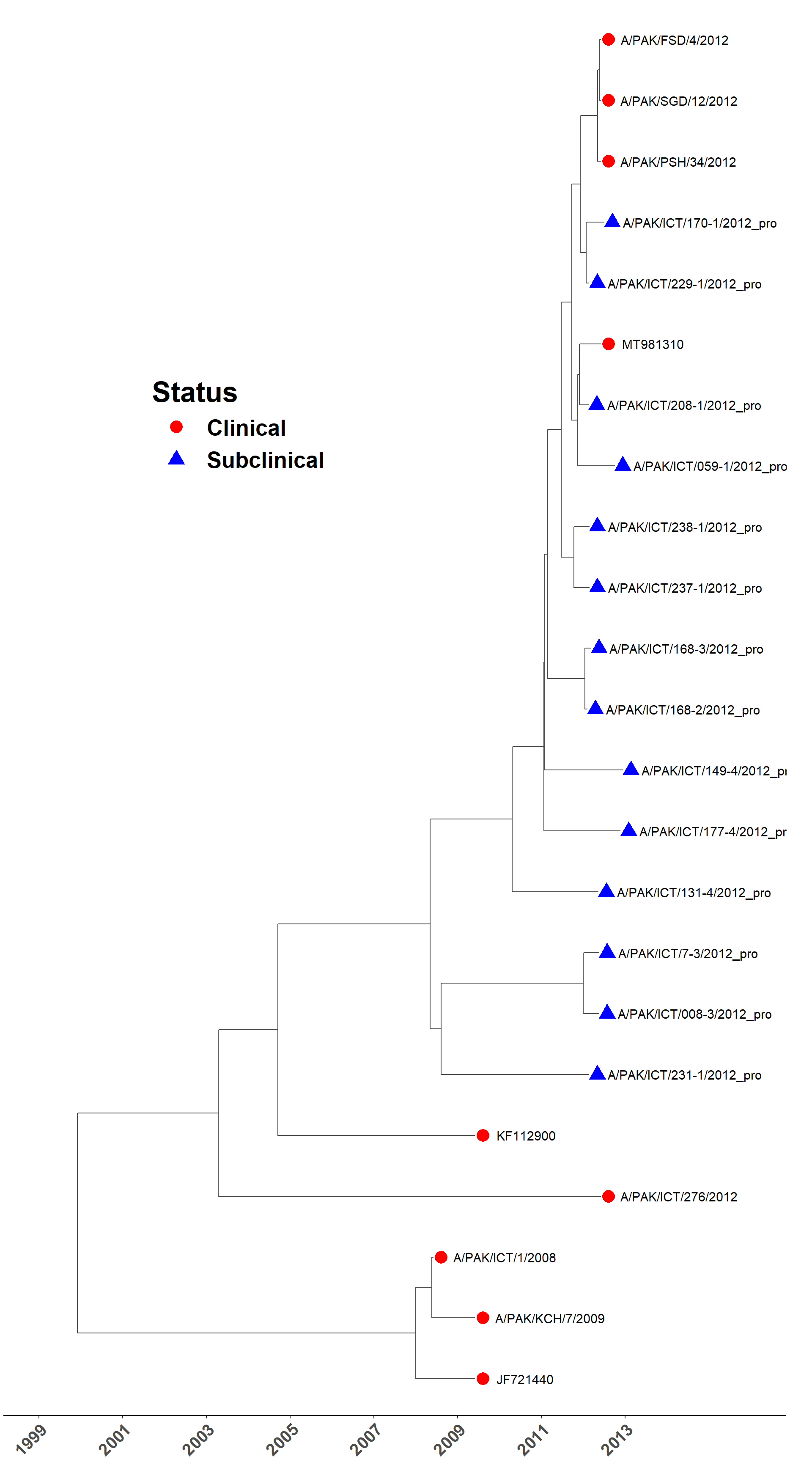
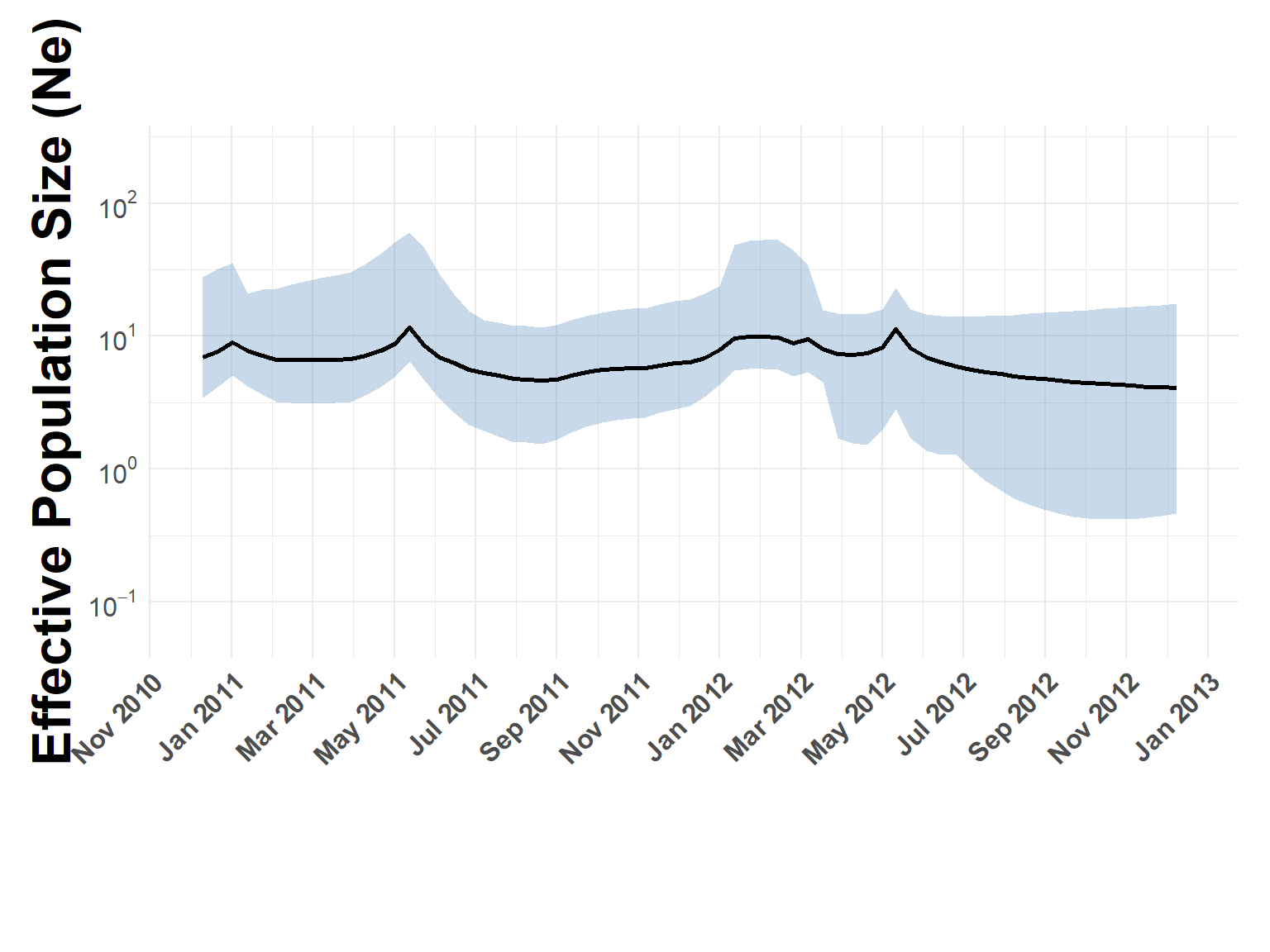
FMDV Effective Population Size
Trees were run using several different clock and prior choices, all showed flat-line Ne. The one here was the simplest, coalescent with contant population size.
Hide code
phylodynamic_process(sero_A.tree, check_stats, x_limits = c("2011-01-01", "2013-01-01"))
*** inla.core.safe: rerun to try to solve negative eigenvalue(s) in the Hessian 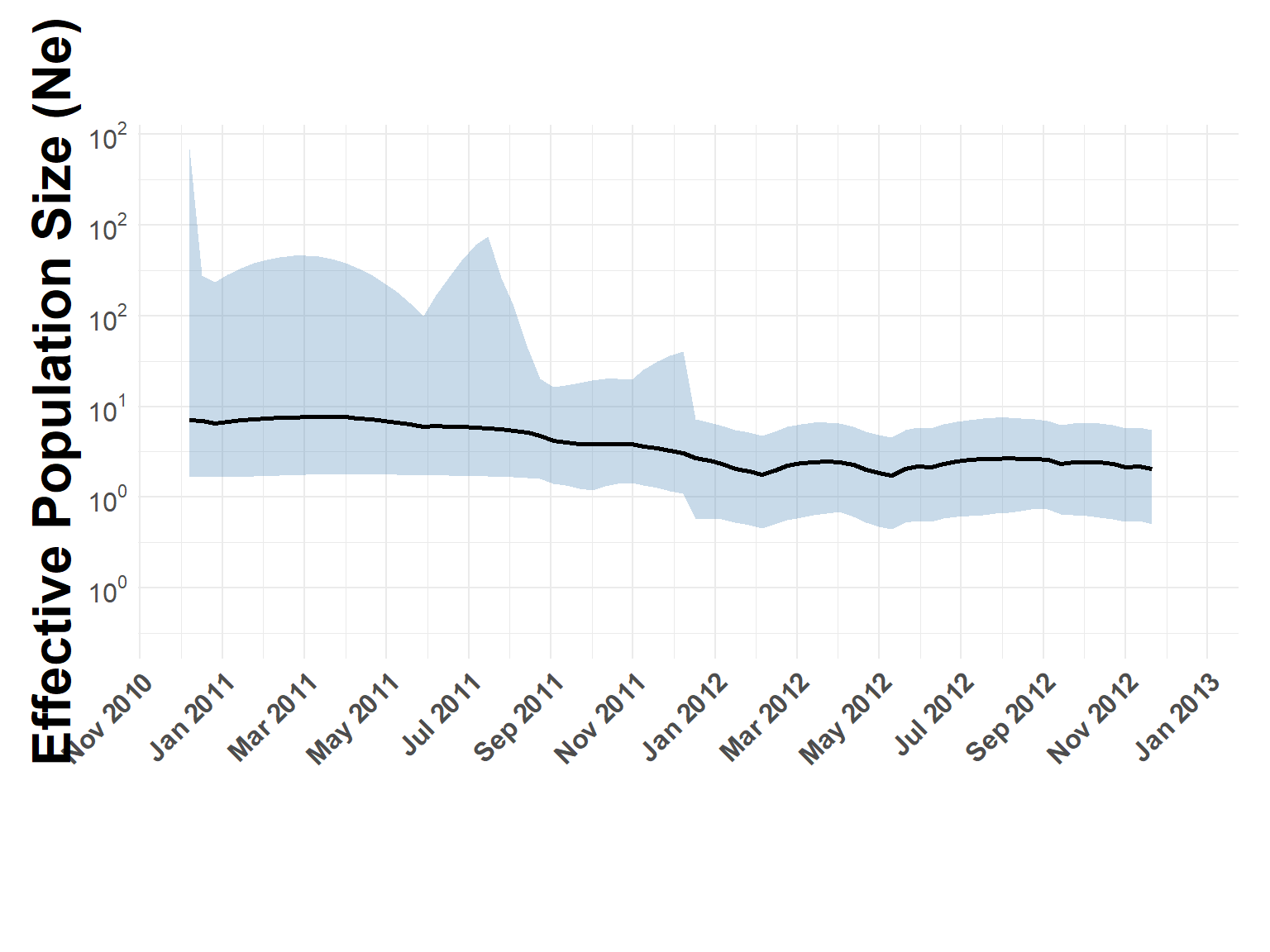
Serotype Asia1
Hide code
check_stats <- get_tracer_stats(here("local/beast/asia1_1/sero_asia1.log.txt"))
keep_stats <- c("joint", "prior", "likelihood", "treeModel.rootHeight", "age.root.",
"treeLength", "tmrca.fmd_a_rev.", "clock.rate")
check_stats %>%
filter(Parameter %in% keep_stats) %>%
gt() %>%
tab_header(
title = md("Serotype Asia1 Stats")) %>%
cols_width(starts_with("Parameter") ~ px(200),
starts_with("label") ~ px(60),
everything() ~ px(95)) %>%
tab_options(table.font.size = "small",
row_group.font.size = "small",
stub.font.size = "small",
column_labels.font.size = "medium",
heading.title.font.size = "large",
data_row.padding = px(2),
heading.title.font.weight = "bold",
column_labels.font.weight = "bold") %>%
opt_stylize(style = 6, color = 'gray')| Serotype Asia1 Stats | |||||
| Parameter | Mean | Median | Q_0.025 | Q_0.975 | ESS |
|---|---|---|---|---|---|
Load MCC tree
Hide code
sero_Asia1.tree <- read.nexus(here("local/beast/asia1_1/sero_asia1.mcc.tre"))
plot_time_tree(sero_Asia1.tree, check_stats, legend_pos = c(0.2, 0.5))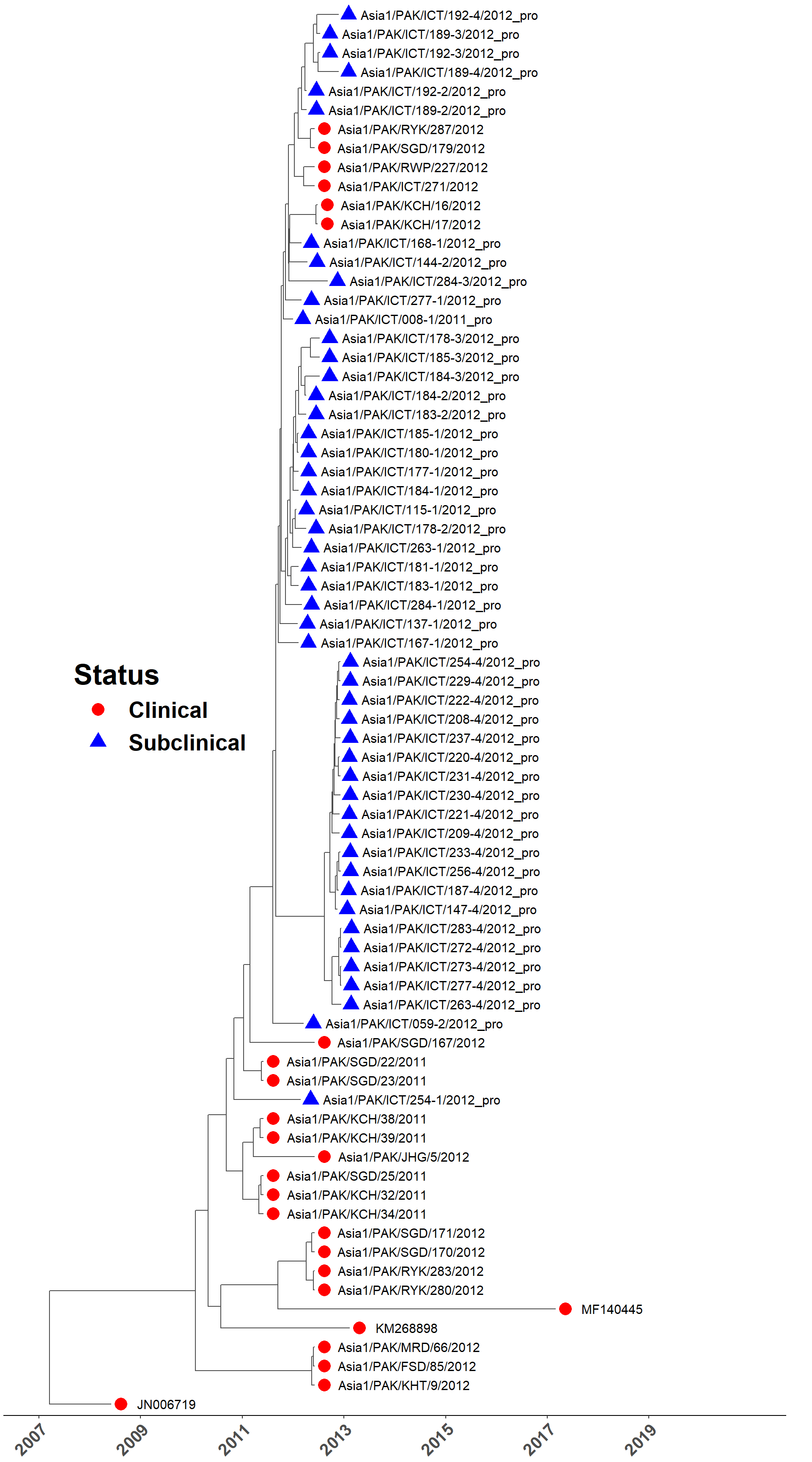
FMDV Effective Population Size
Flat Ne, like the other serotypes
Hide code
phylodynamic_process(sero_Asia1.tree, check_stats, x_limits = c("2011-01-01", "2013-01-01"))
*** inla.core.safe: rerun to try to solve negative eigenvalue(s) in the Hessian 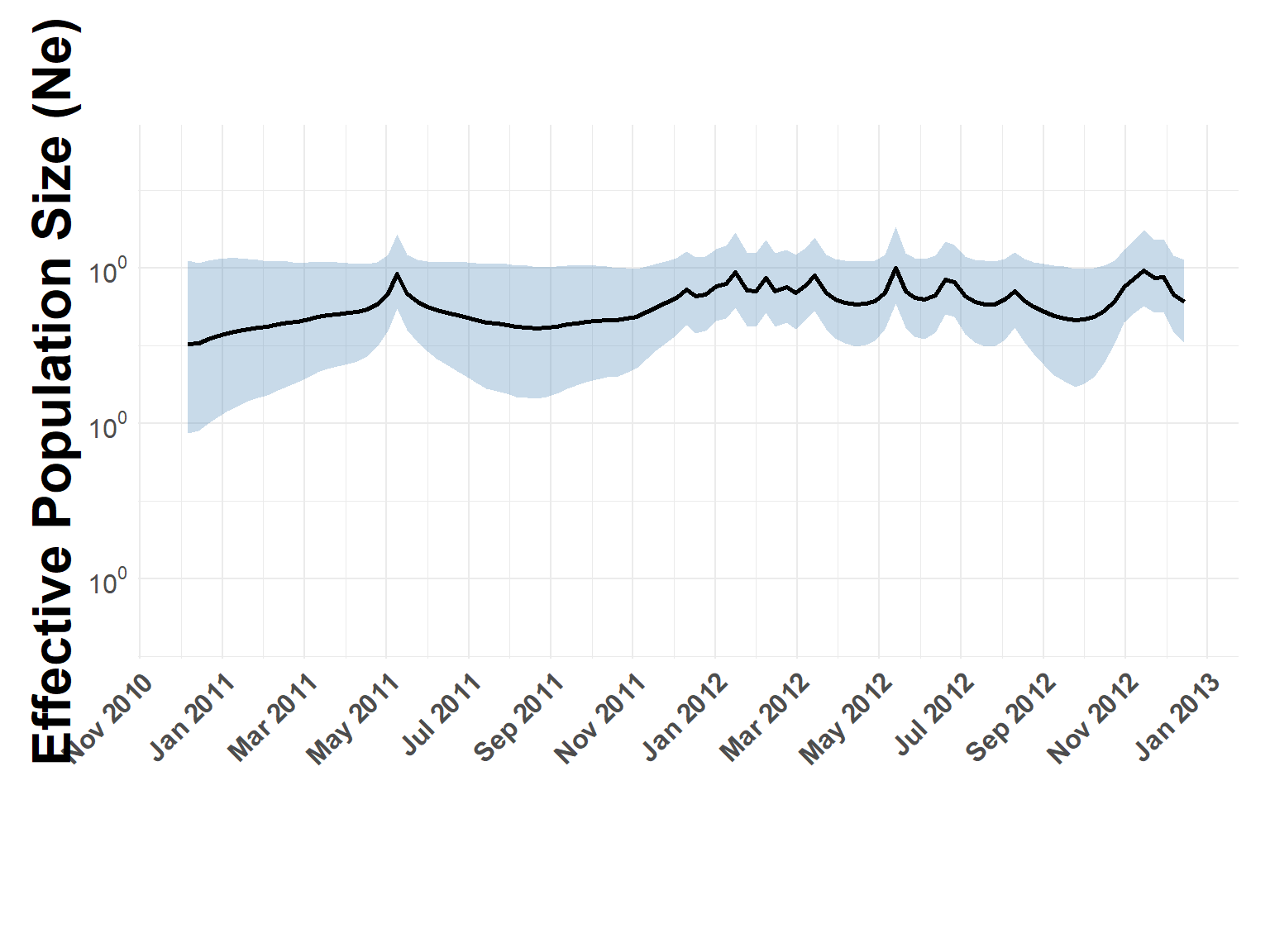
Serotype O
Hide code
check_stats <- get_tracer_stats(here("local/beast/o_1/sero_o.log.txt"))
keep_stats <- c("joint", "prior", "likelihood", "treeModel.rootHeight", "age.root.",
"treeLength", "tmrca.fmd_a_rev.", "clock.rate")
check_stats %>%
filter(Parameter %in% keep_stats) %>%
gt() %>%
tab_header(
title = md("Serotype O Stats")) %>%
cols_width(starts_with("Parameter") ~ px(200),
starts_with("label") ~ px(60),
everything() ~ px(95)) %>%
tab_options(table.font.size = "small",
row_group.font.size = "small",
stub.font.size = "small",
column_labels.font.size = "medium",
heading.title.font.size = "large",
data_row.padding = px(2),
heading.title.font.weight = "bold",
column_labels.font.weight = "bold") %>%
opt_stylize(style = 6, color = 'gray')| Serotype O Stats | |||||
| Parameter | Mean | Median | Q_0.025 | Q_0.975 | ESS |
|---|---|---|---|---|---|
Load MCC tree
Hide code
sero_O.tree <- read.nexus(here("local/beast/o_1/sero_o.mcc.tre"))
plot_time_tree(sero_O.tree, check_stats, legend_pos = c(0.2, 0.5))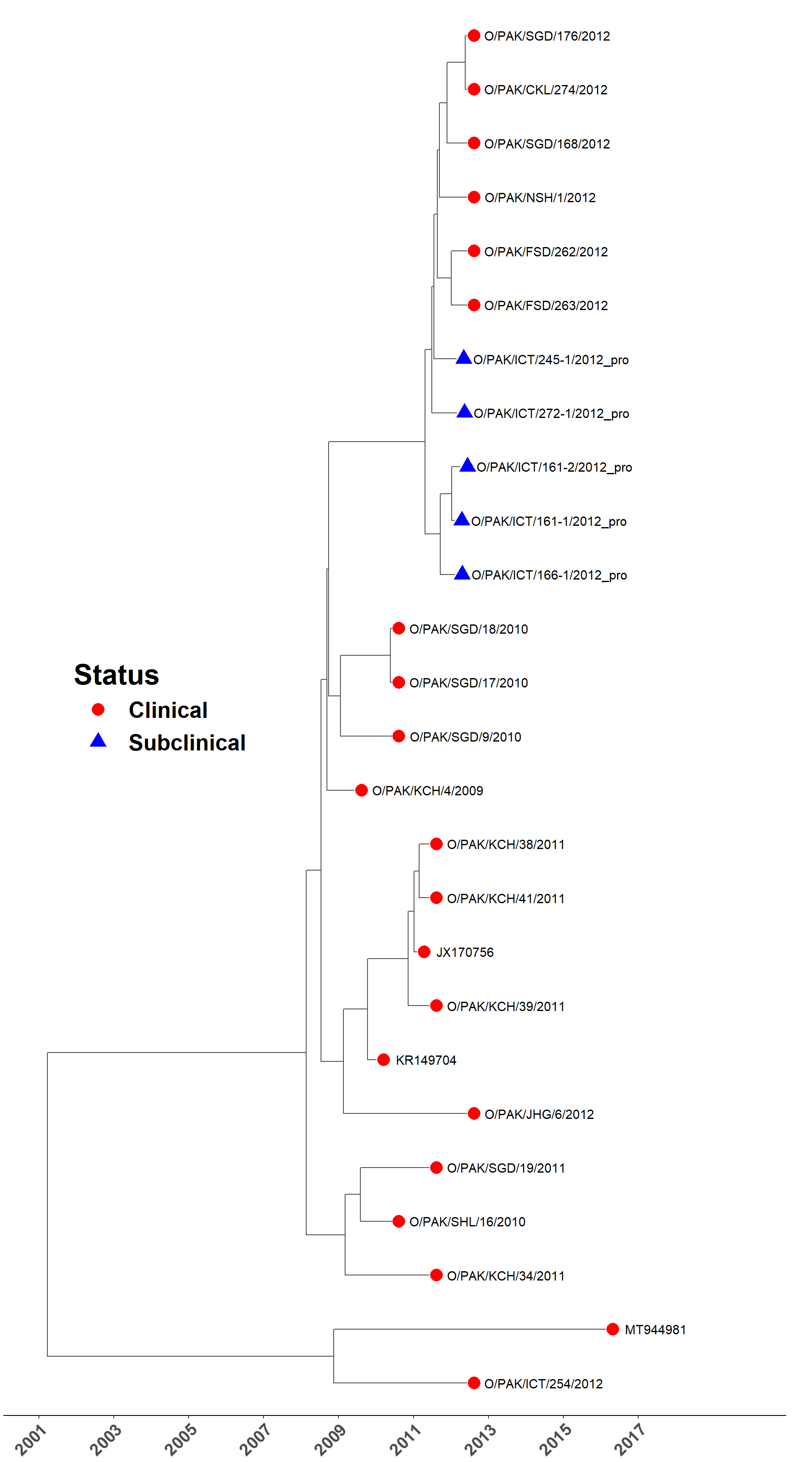
FMDV Effective Population Size
Flat Ne, like the other serotypes
Hide code
phylodynamic_process(sero_O.tree, check_stats, x_limits = c("2011-01-01", "2013-01-01"))
*** inla.core.safe: rerun to try to solve negative eigenvalue(s) in the Hessian