Entanglement
Assessing recombination history through comparison of subgenomic region phylogenies
Read Gene Trees
Segment specific phylogenies. Note that the_rev subfix indicates that the original Nexus files were revised to swap tip labels for names in the description block (see, preprocessing)
Show code
# phylogeny list from directory
tree_files <- list.files(here("local/paktrees/original"), pattern="_rev\\.nex$")
# read ML trees & convert
IRES_Lpro.tree <- read.nexus(here("local/paktrees/original", tree_files[8]))
IRES_Lpro.den <- get_dendro(
extract_subtree(IRES_Lpro.tree))
seg_2C.tree <- read.nexus(here("local/paktrees/original", tree_files[1]))
seg_2C.den <- get_dendro(
extract_subtree(seg_2C.tree))
seg_3A.tree <- read.nexus(here("local/paktrees/original", tree_files[2]))
seg_3A.den <- get_dendro(
extract_subtree(seg_3A.tree))
seg_3C.tree <- read.nexus(here("local/paktrees/original", tree_files[3]))
seg_3C.den <- get_dendro(
extract_subtree(seg_3C.tree))
seg_3D.tree <- read.nexus(here("local/paktrees/original", tree_files[4]))
seg_3D.den <- get_dendro(
extract_subtree(seg_3D.tree))Entanglement
Entanglement scores range from 0.00 - 1.00 with a values of 0.00 indicating the trees are exactly the same and a score of 1.00 indicating they are opposites. Essentially, the score can be interpreted as a proportion or percent change.
Table to summarize comparisons
Show code
entangle_tab <- as.data.frame(matrix(nrow = 10, ncol = 2))
names(entangle_tab) <- c("Compared", "Score")Select tabs to view comparisons
Show code
dends <- dendlist(IRES_Lpro.den, seg_2C.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[1] <- "IRES_Lpro vs. 2C"
entangle_tab$Score[1] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "IRES_Lpro",
cex_main_left = 4,
main_right = "2C",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)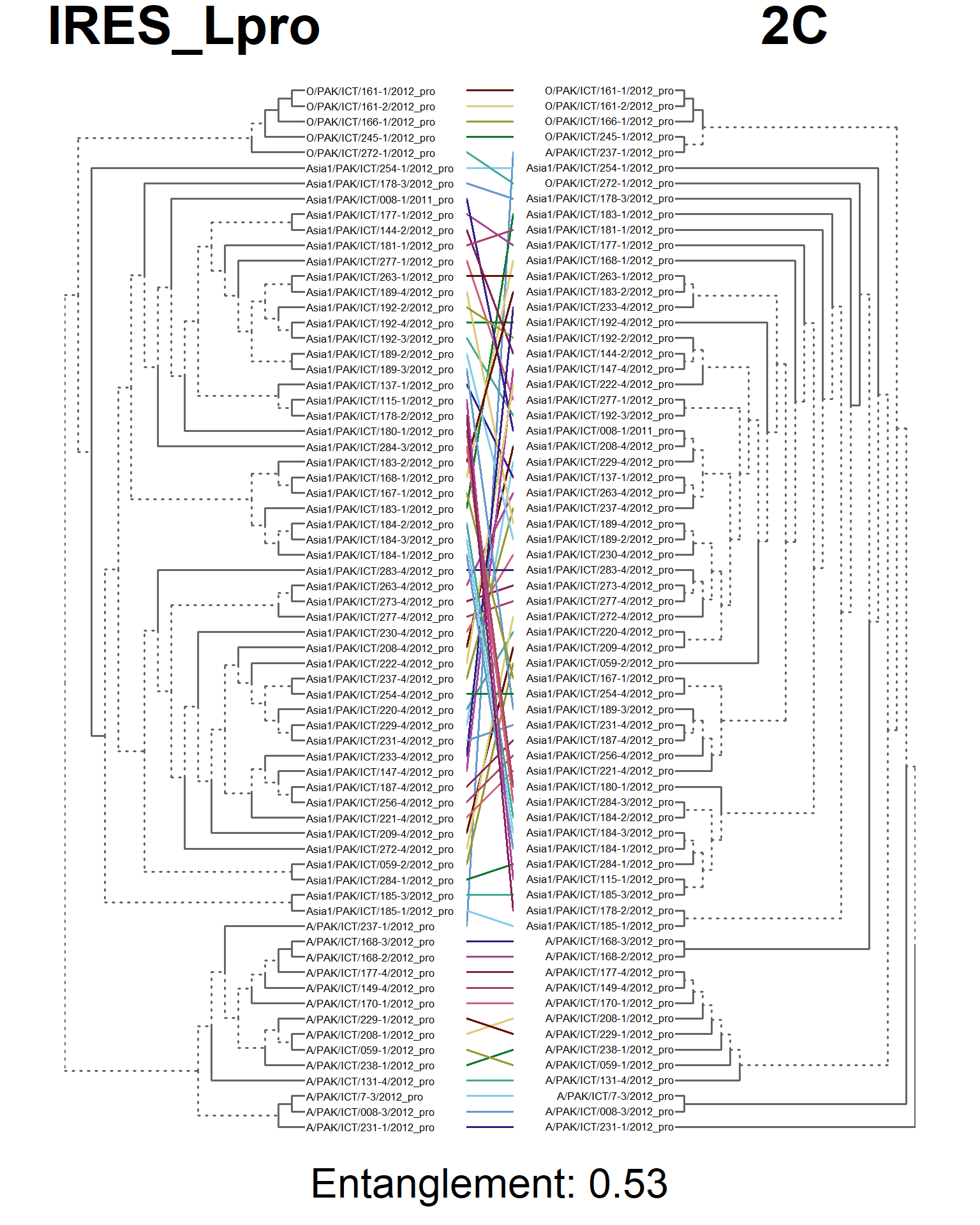
Show code
dends <- dendlist(IRES_Lpro.den, seg_3A.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[2] <- "IRES_Lpro vs. 3A"
entangle_tab$Score[2] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "IRES_Lpro",
cex_main_left = 4,
main_right = "3A",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)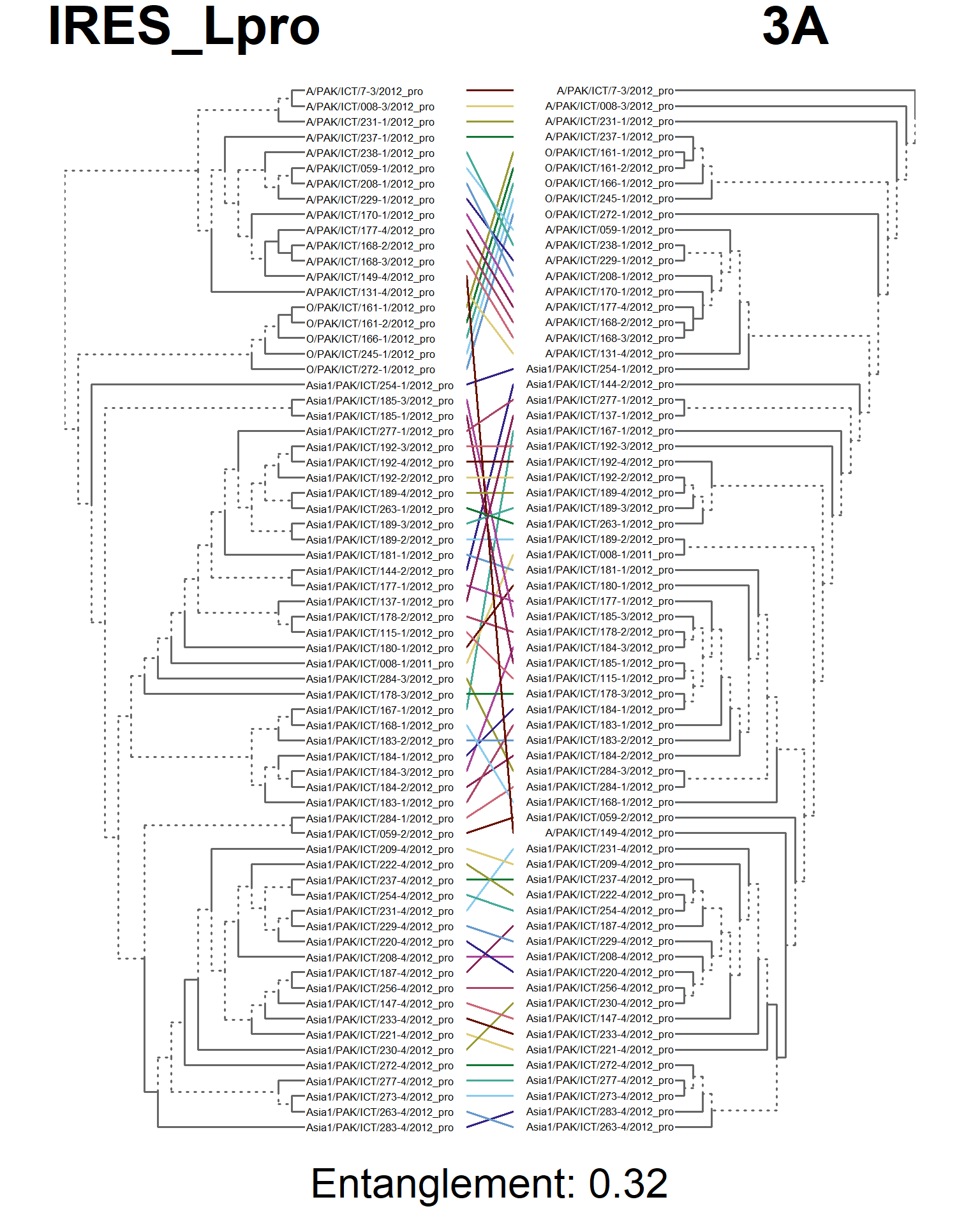
Show code
dends <- dendlist(IRES_Lpro.den, seg_3C.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[3] <- "IRES_Lpro vs. 3C"
entangle_tab$Score[3] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "IRES_Lpro",
cex_main_left = 4,
main_right = "3C",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)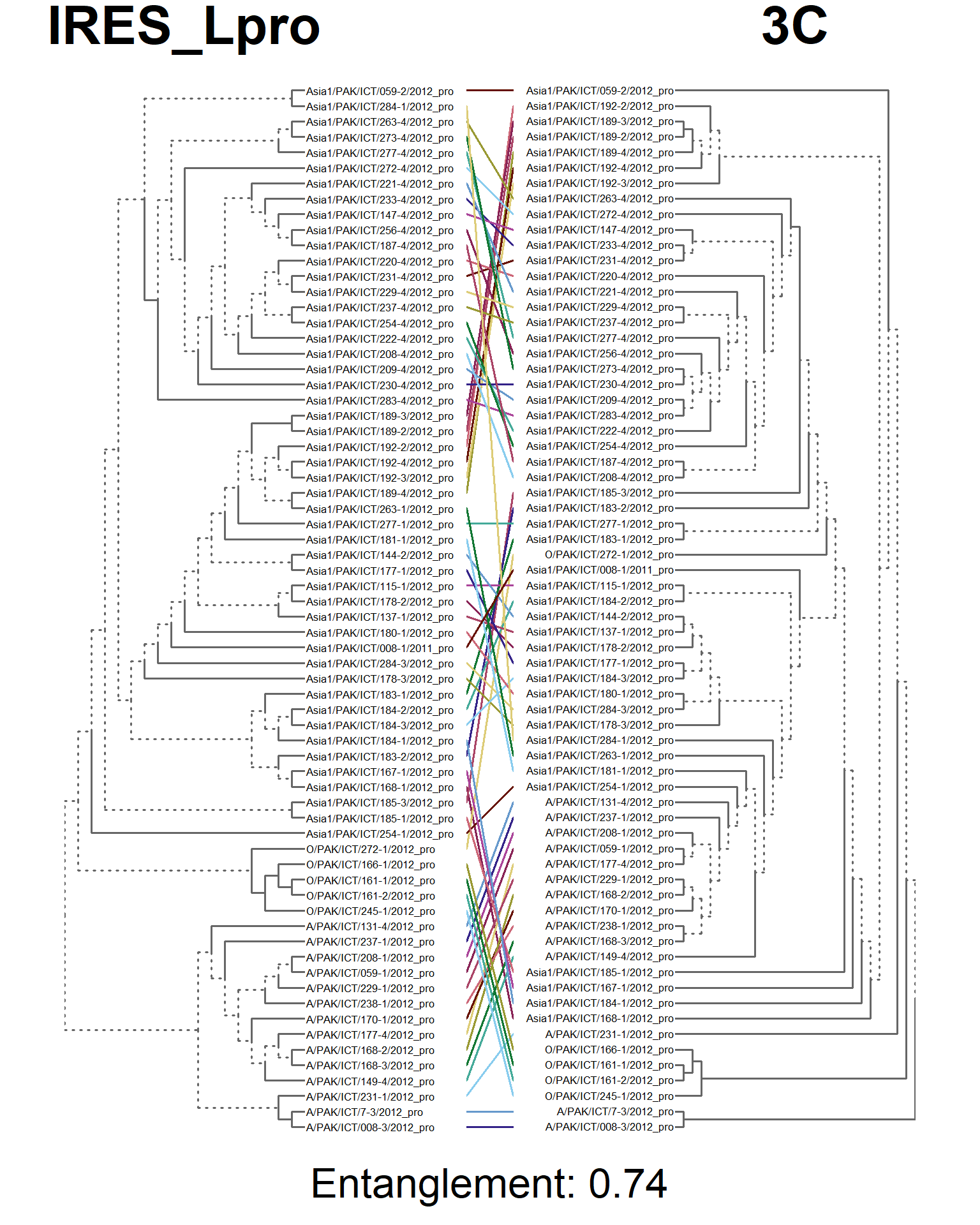
Show code
dends <- dendlist(IRES_Lpro.den, seg_3D.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[4] <- "IRES_Lpro vs. 3D"
entangle_tab$Score[4] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "IRES_Lpro",
cex_main_left = 4,
main_right = "3D",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)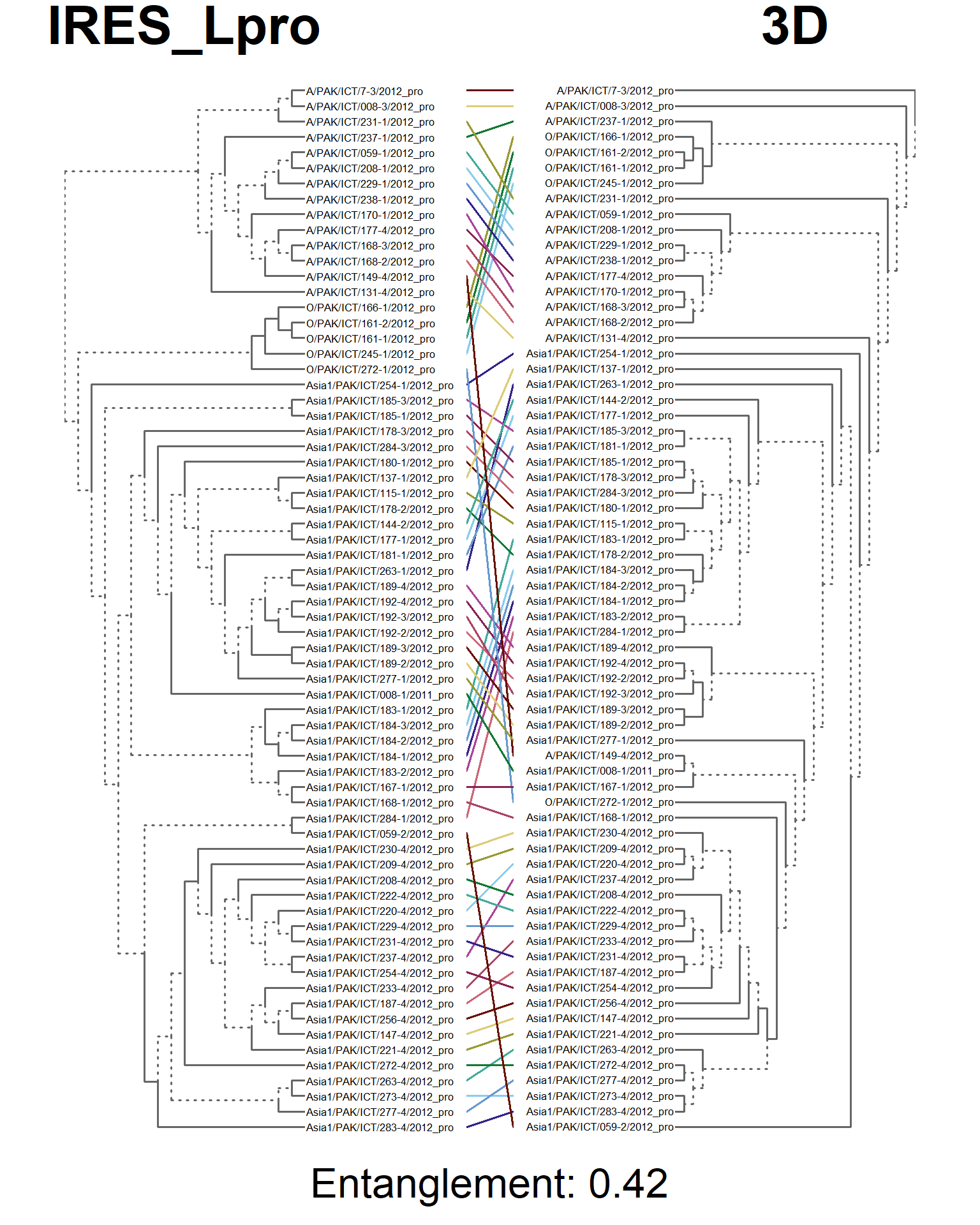
Show code
dends <- dendlist(seg_2C.den, seg_3A.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[5] <- "2C vs. 3A"
entangle_tab$Score[5] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "2C",
cex_main_left = 4,
main_right = "3A",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)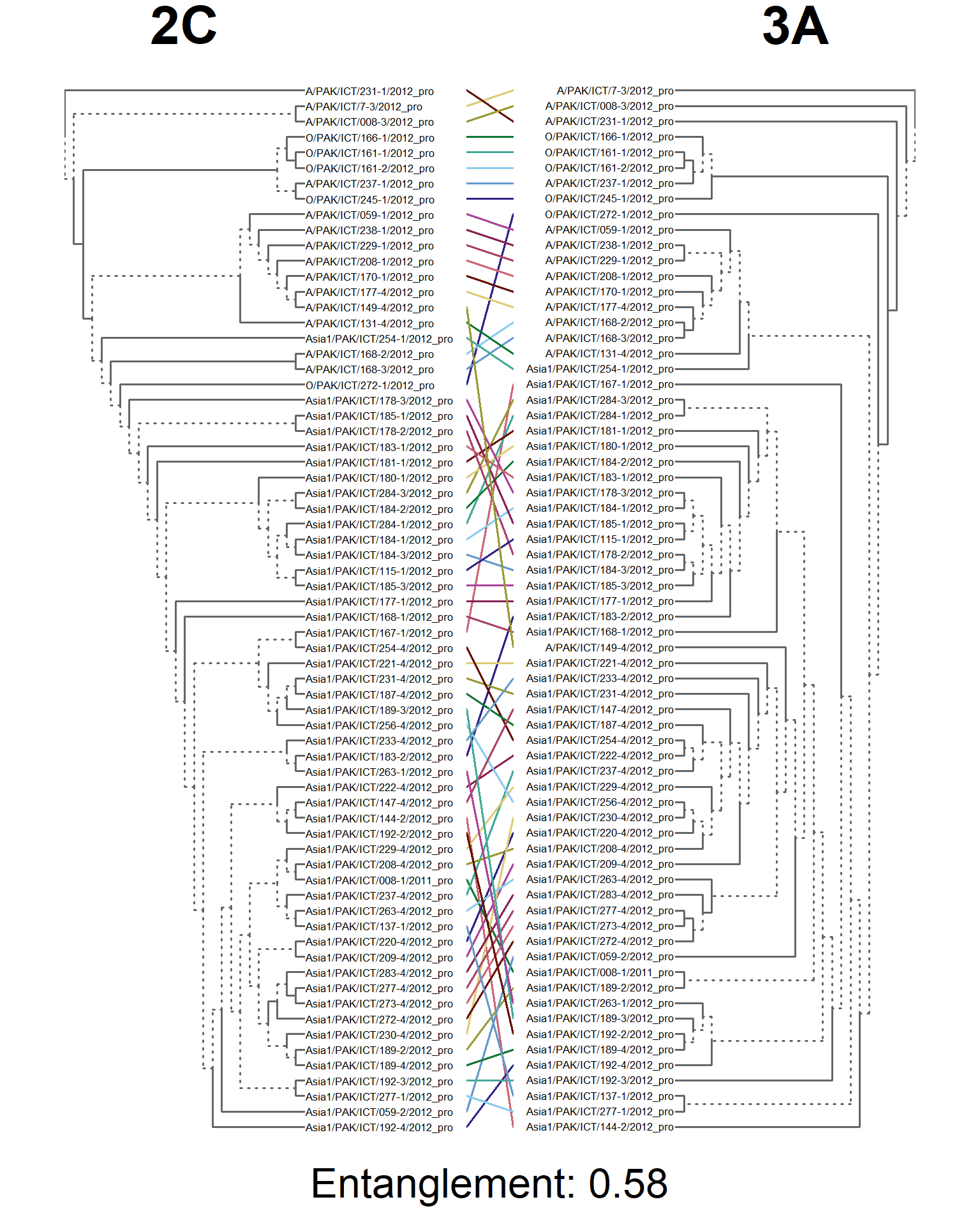
Show code
dends <- dendlist(seg_2C.den, seg_3C.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[6] <- "2C vs. 3C"
entangle_tab$Score[6] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "2C",
cex_main_left = 4,
main_right = "3C",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)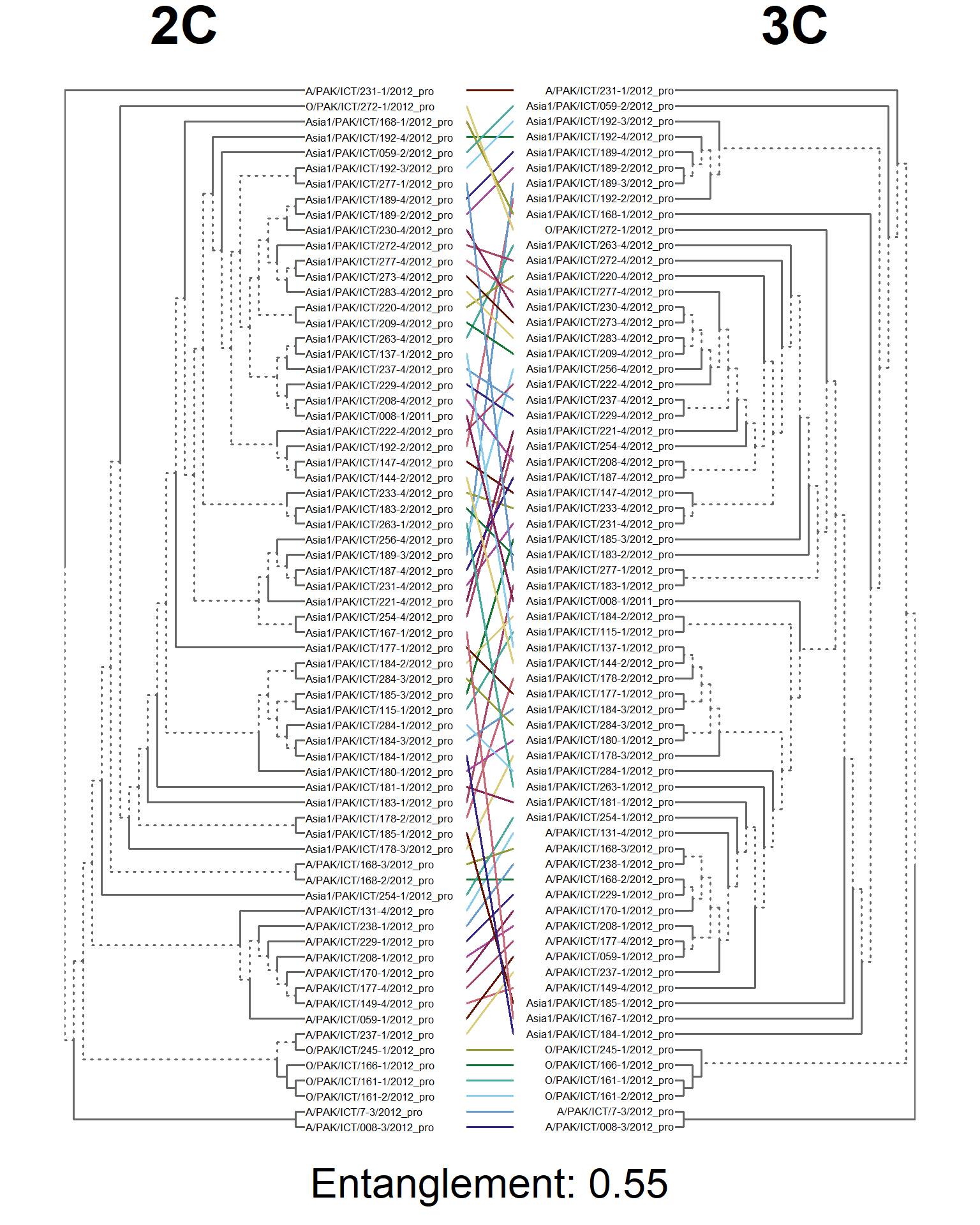
Show code
dends <- dendlist(seg_2C.den, seg_3D.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[7] <- "2C vs. 3D"
entangle_tab$Score[7] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "2C",
cex_main_left = 4,
main_right = "3D",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)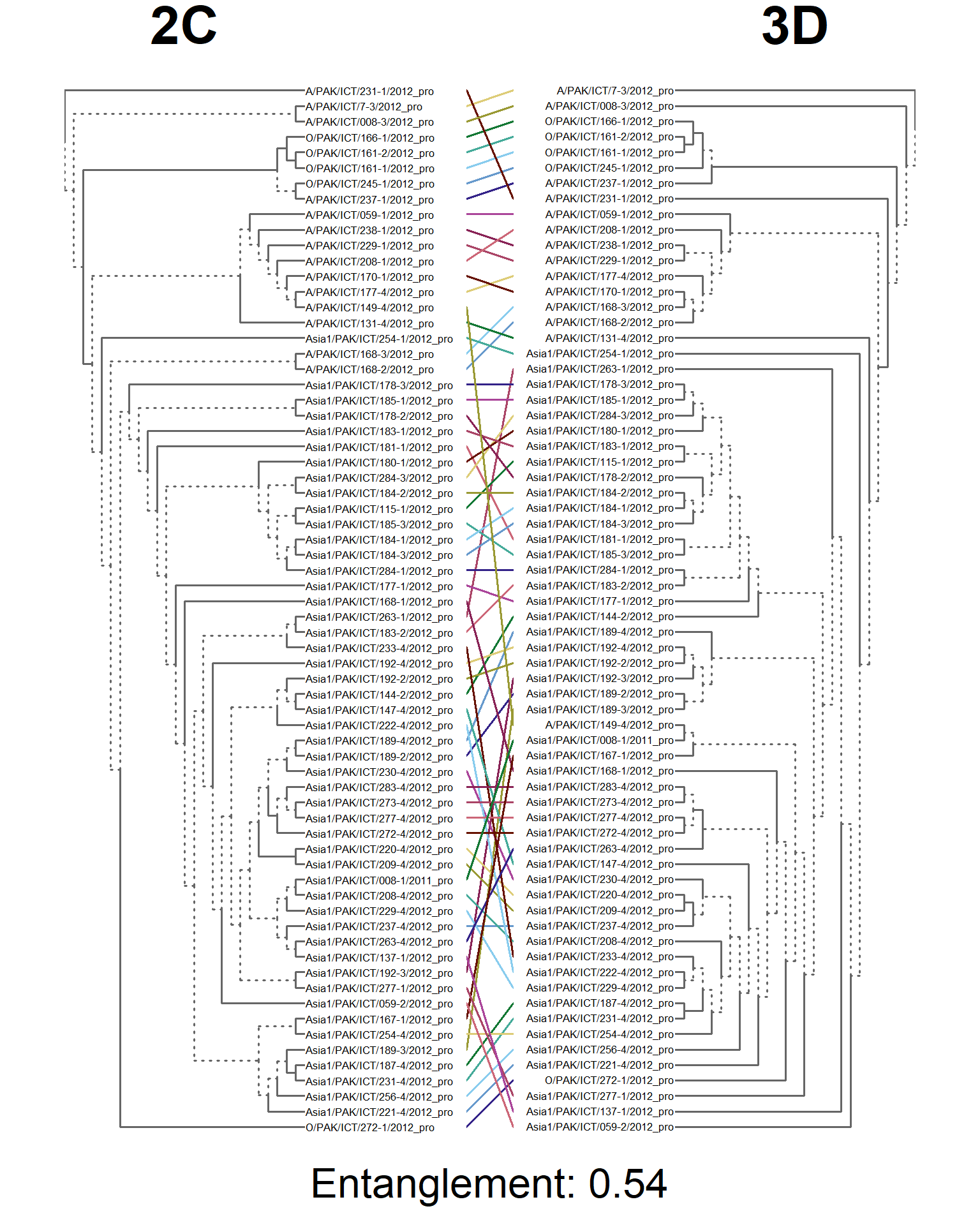
Show code
dends <- dendlist(seg_3A.den, seg_3C.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[8] <- "3A vs. 3C"
entangle_tab$Score[8] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "3A",
cex_main_left = 4,
main_right = "3C",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)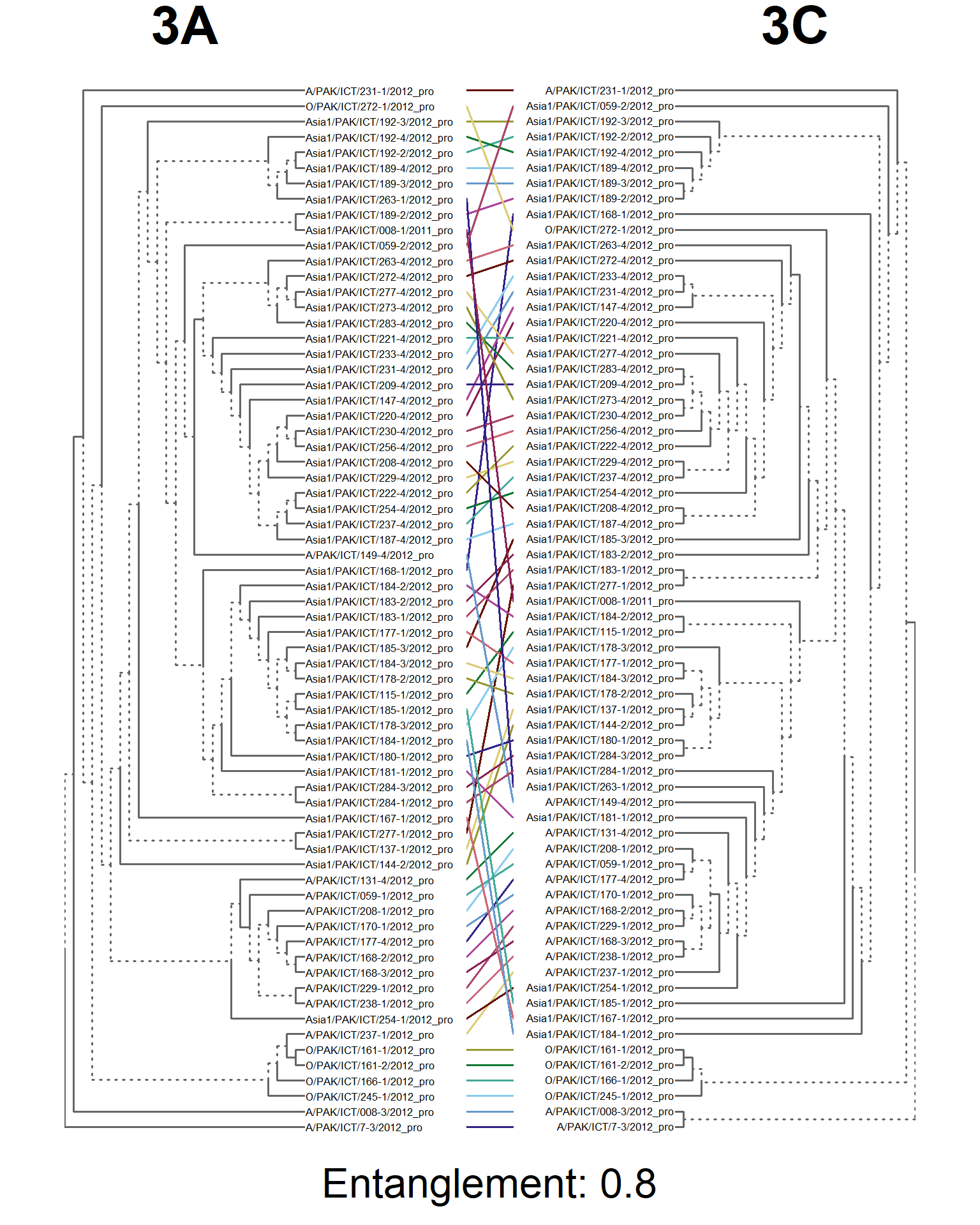
Show code
dends <- dendlist(seg_3A.den, seg_3D.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[9] <- "3A vs. 3D"
entangle_tab$Score[9] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "3A",
cex_main_left = 4,
main_right = "3D",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)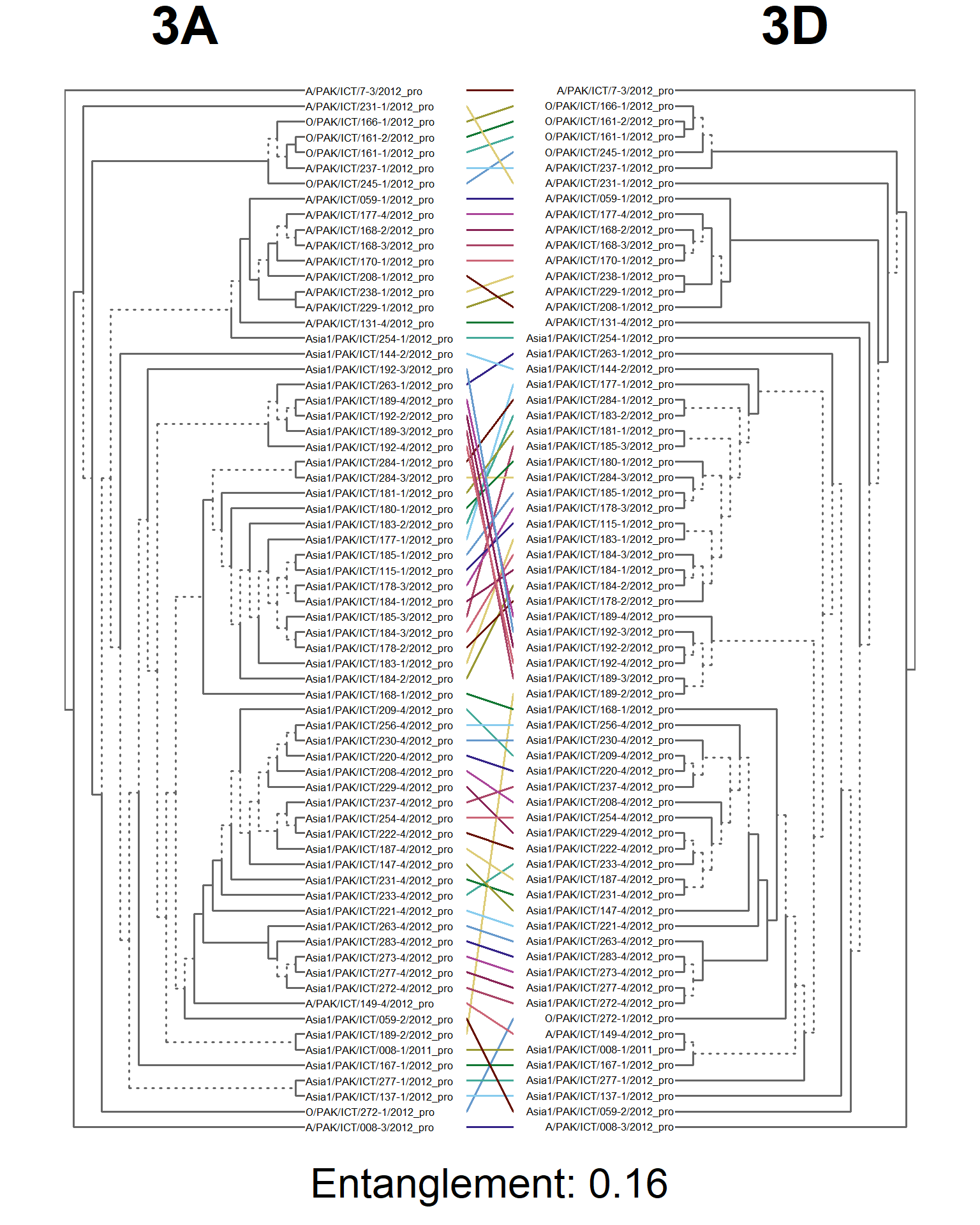
Show code
dends <- dendlist(seg_3C.den, seg_3D.den)
entang_score <- round(entanglement(dends), 2)
entangle_tab$Compared[10] <- "3C vs. 3D"
entangle_tab$Score[10] <- entang_score
x <- dends %>% dendextend::untangle(method = "step2side")
x %>% set("rank_branches") %>%
set("branches_lwd", 1.5) %>%
set("branches_col", value = c("gray40")) %>%
tanglegram(main_left = "3C",
cex_main_left = 4,
main_right = "3D",
cex_main_right = 4,
columns_width = c(5,0.5,5),
lwd = 1.8,
lab.cex = 0.8,
dLeaf_left = -0.15,
dLeaf_right = 0.15,
margin_outer = 4,
margin_inner = 10,
color_lines = tol(12),
axes=FALSE,
sub = paste("Entanglement:", entang_score),
common_subtrees_color_branches = FALSE)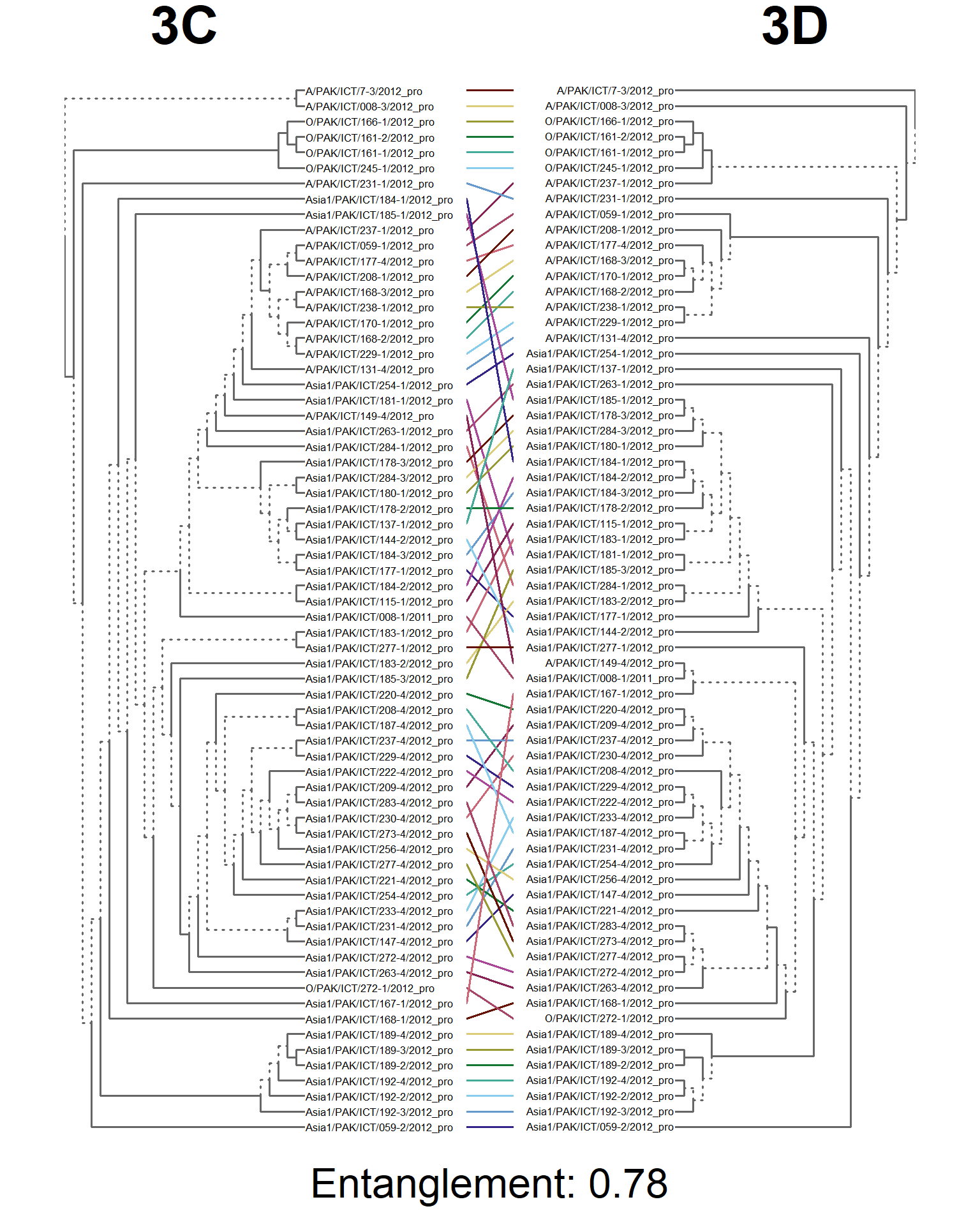
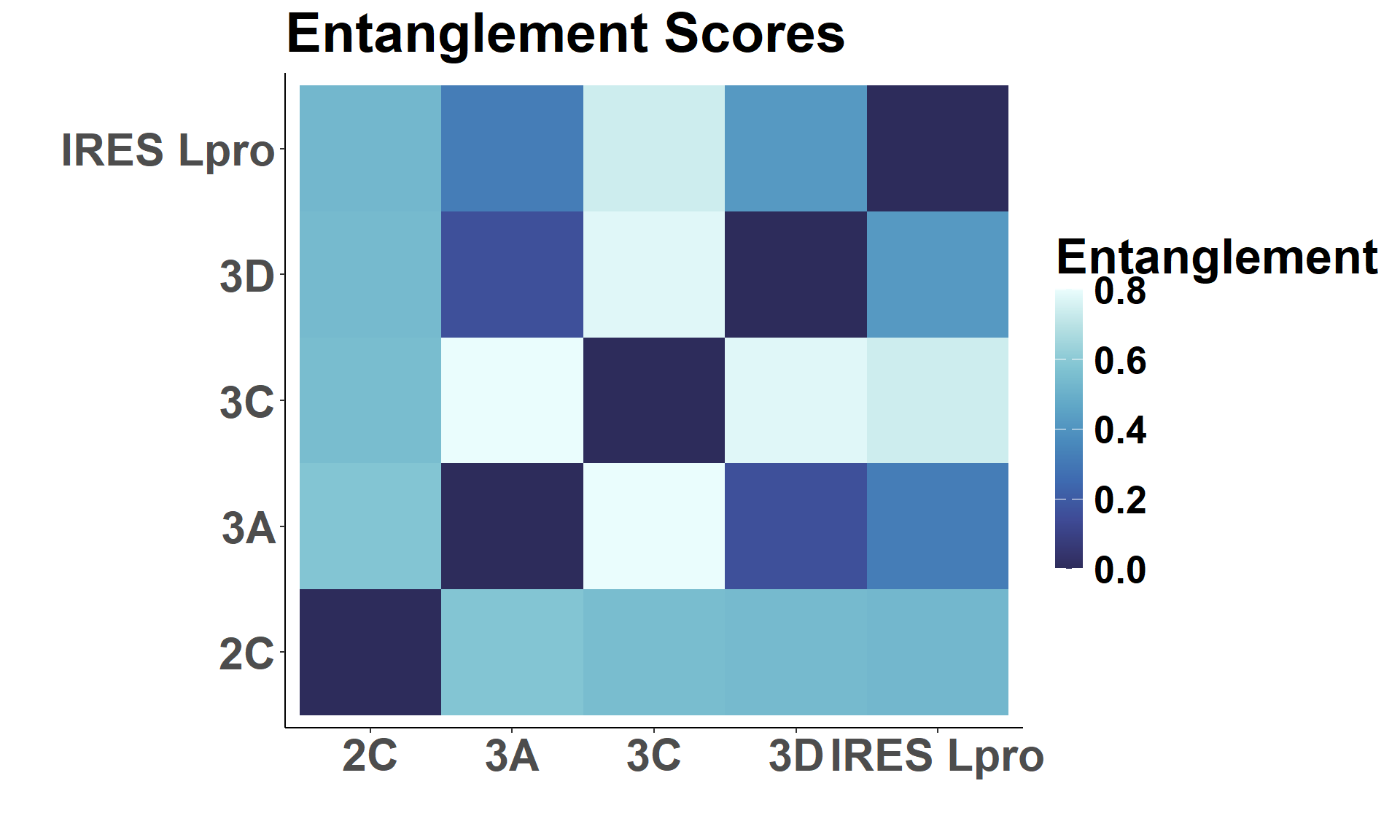
Entanglement Scores
Show code
entangle_tab %>%
gt() %>%
tab_header(
title = md("Score Summary")) %>%
cols_width(everything() ~ px(150)) %>%
tab_options(table.font.size = "small",
row_group.font.size = "small",
stub.font.size = "small",
column_labels.font.size = "medium",
heading.title.font.size = "large",
data_row.padding = px(2),
heading.title.font.weight = "bold",
column_labels.font.weight = "bold") %>%
opt_stylize(style = 6, color = 'gray')| Score Summary | |
| Compared | Score |
|---|---|
Matrix View
Show code
mat_view <- entangle_tab %>%
separate(Compared, into = c("Var1", "Var2"), sep = " vs. ", remove = FALSE)
mat_view <- mat_view %>%
select(Var1, Var2, Score)
unique_vars <- unique(c(mat_view$Var1, mat_view$Var2))
self_comparisons <- data.frame(
Var1 = unique_vars,
Var2 = unique_vars,
Score = 0 # self-comparisons
)
mat_view <- bind_rows(mat_view, self_comparisons)
mat_view <- bind_rows(
mat_view,
mat_view %>% rename(Var1 = Var2, Var2 = Var1)
)
mat_view <- mat_view %>%
distinct()
mat_view$Var1 <- gsub("_", " ", mat_view$Var1)
mat_view$Var2 <- gsub("_", " ", mat_view$Var2)
ggplot(mat_view, aes(Var1, Var2, fill = Score)) +
geom_tile() +
scale_fill_gradientn(
colors = (pals::ocean.ice(100)[20:100])
) +
theme_classic() +
theme(
plot.margin = unit(c(2, 2, 2, 2), "mm"),
axis.title.x = element_text(size = 24, face = "bold"),
axis.title.y = element_text(size = 24, face = "bold"),
axis.text.x = element_text(size = 23, face = "bold"),
axis.text.y = element_text(size = 23, face = "bold"),
legend.direction = "vertical",
legend.position = "right",
strip.text = element_blank(),
strip.background = element_blank(),
legend.key.size = unit(2, "line"),
legend.key.width = unit(1, "line"),
legend.text = element_text(size = 20, face = "bold"),
legend.title = element_text(size = 25, face = "bold"),
plot.title = element_text(size = 28, face = "bold")
) +
labs(title = "Entanglement Scores",
x = " ", y = " ", fill = "Entanglement")