Longitudinal Analysis
Mixed effect models for individual, time-dependent observations
Organize Data
Hide code
longitud_df <- antem_df %>%
filter(group != "Group 1") %>%
mutate(y_shed = if_else(nasal == 0, NA, nasal),
y_shed = if_else(nasal == 45, 0, y_shed),
y_serum = if_else(serum == 0, NA, serum),
y_serum = if_else(serum == 45, 0, y_serum),
temp_sc = as.numeric(scale(temp, scale = TRUE, center = TRUE)),
time_step = dpe + 1)
#plot(density(longitud_df$y_serum, na.rm=T)) # ~normalShedding Rates
Run model for shedding (nasal swabs)
Hide code
return_quants <- c(0.025, 0.05, 0.25, 0.5, 0.75, 0.95, 0.975)
pc_prec_rw <- list(theta = list(prior="pc.prec",
param=c(1, 0.001)))
pc_prec_iid <- list(theta = list(prior="pc.prec",
param=c(3, 0.001)))
shed_form <- y_shed ~ 1 + f(time_step,
model = "rw1",
constr = TRUE,
scale.model = TRUE,
hyper=pc_prec_rw) +
f(animal,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
f(exp_type,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
temp
shed_model <- inla(shed_form,
data = longitud_df,
family = "gaussian",
quantiles = return_quants,
verbose=FALSE,
control.compute=list(dic = TRUE, cpo = FALSE, waic = TRUE))
# summary(shed_model)Plot Time Effect
Hide code
time_df <- shed_model$summary.random$time_step[, c(1, 4:10)]
names(time_df) <- c("DPE", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
time_df$DPE <- time_df$DPE - 1
plot_time_rw(time_df)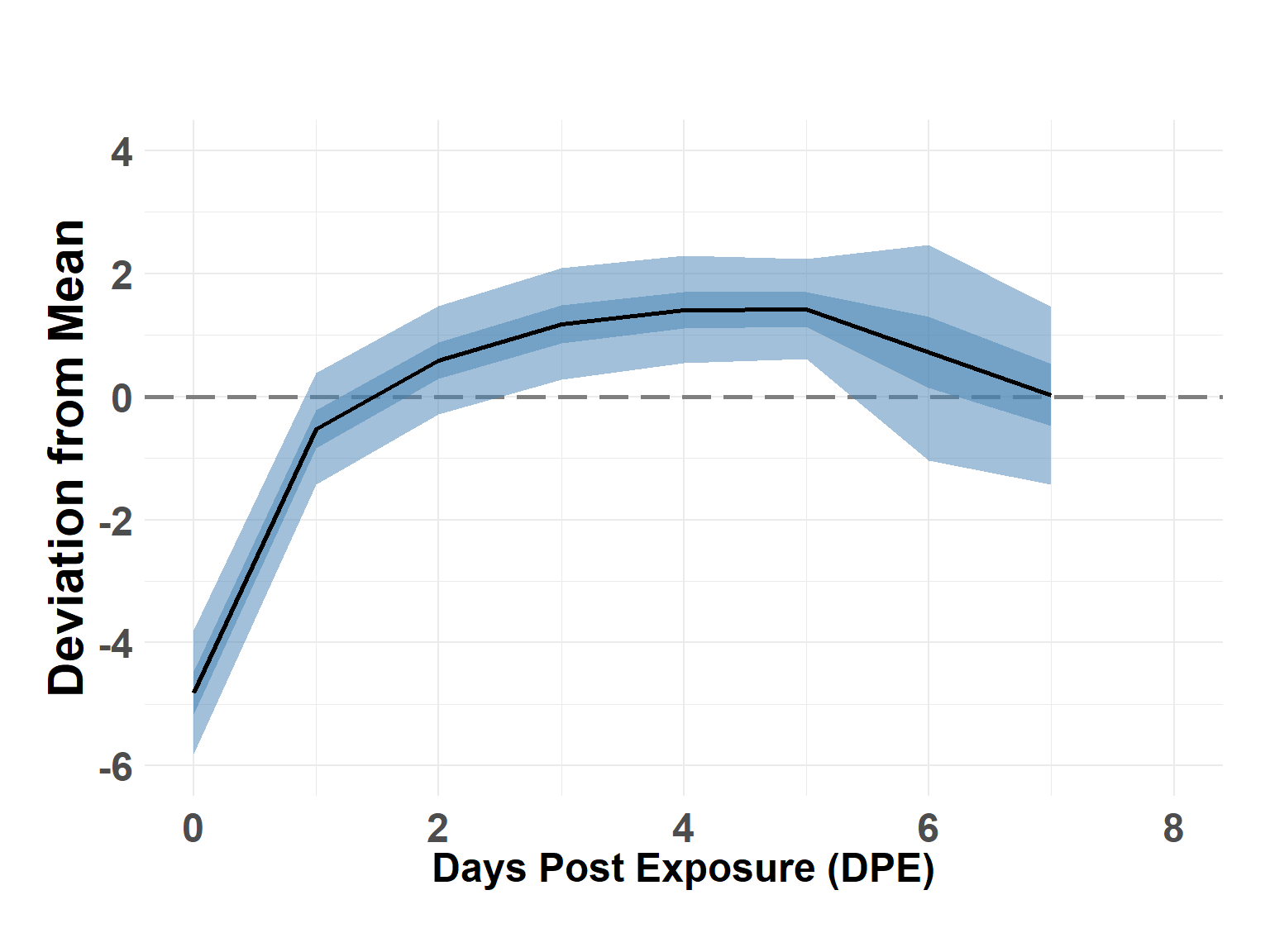
Plot animals
Hide code
animal_df <- shed_model$summary.random$animal[, c(1, 4:10)]
names(animal_df) <- c("Animal", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
plot_animals_linerange(animal_df)Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.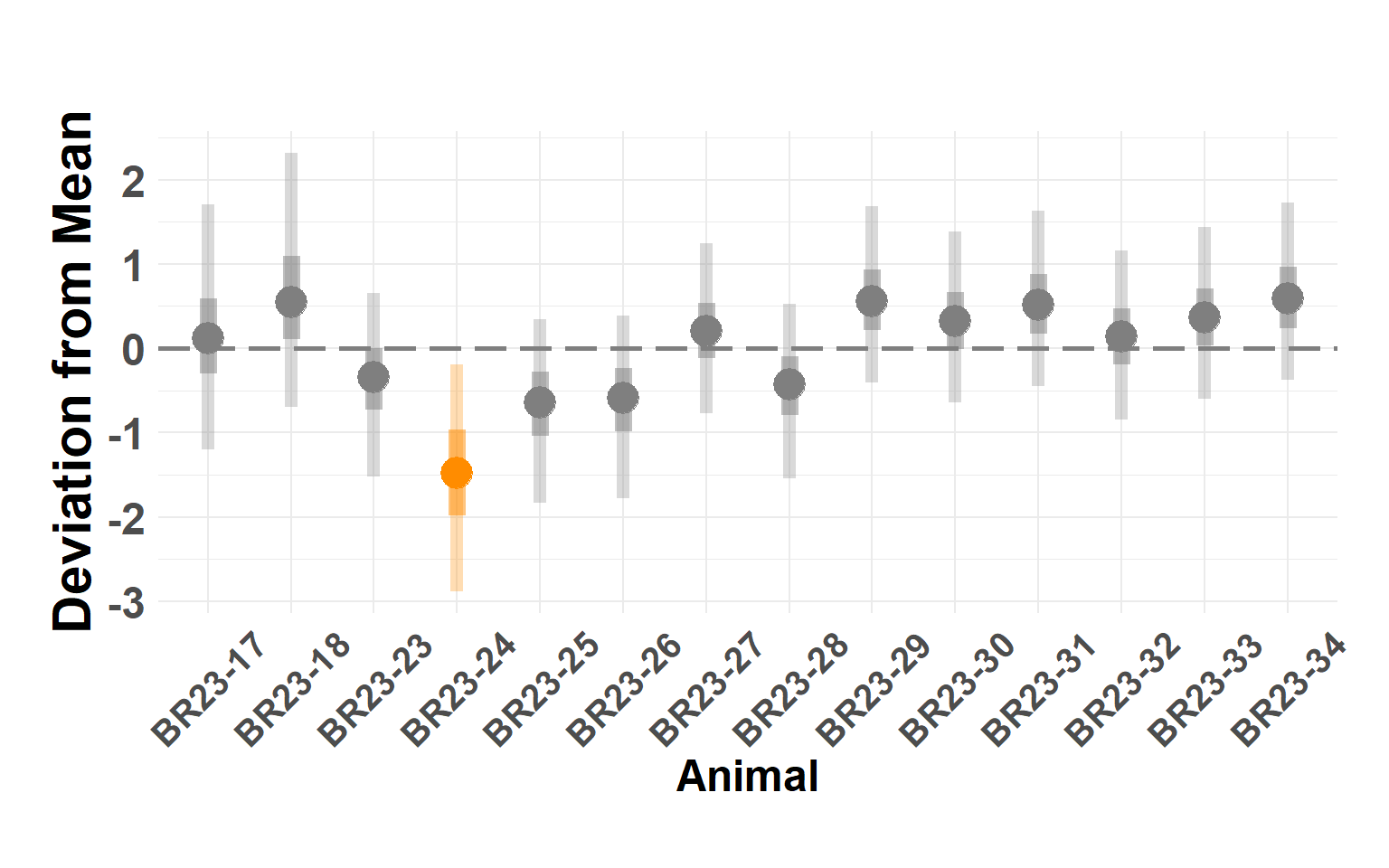
Plot exposure type
Hide code
exptype_df <- shed_model$summary.random$exp_type[, c(1, 4:10)]
names(exptype_df) <- c("type", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
exptype_df$type <- as.factor(exptype_df$type)
levels(exptype_df$type) <- c("Contact", "Inoculation")
plot_exposure_linerange(exptype_df)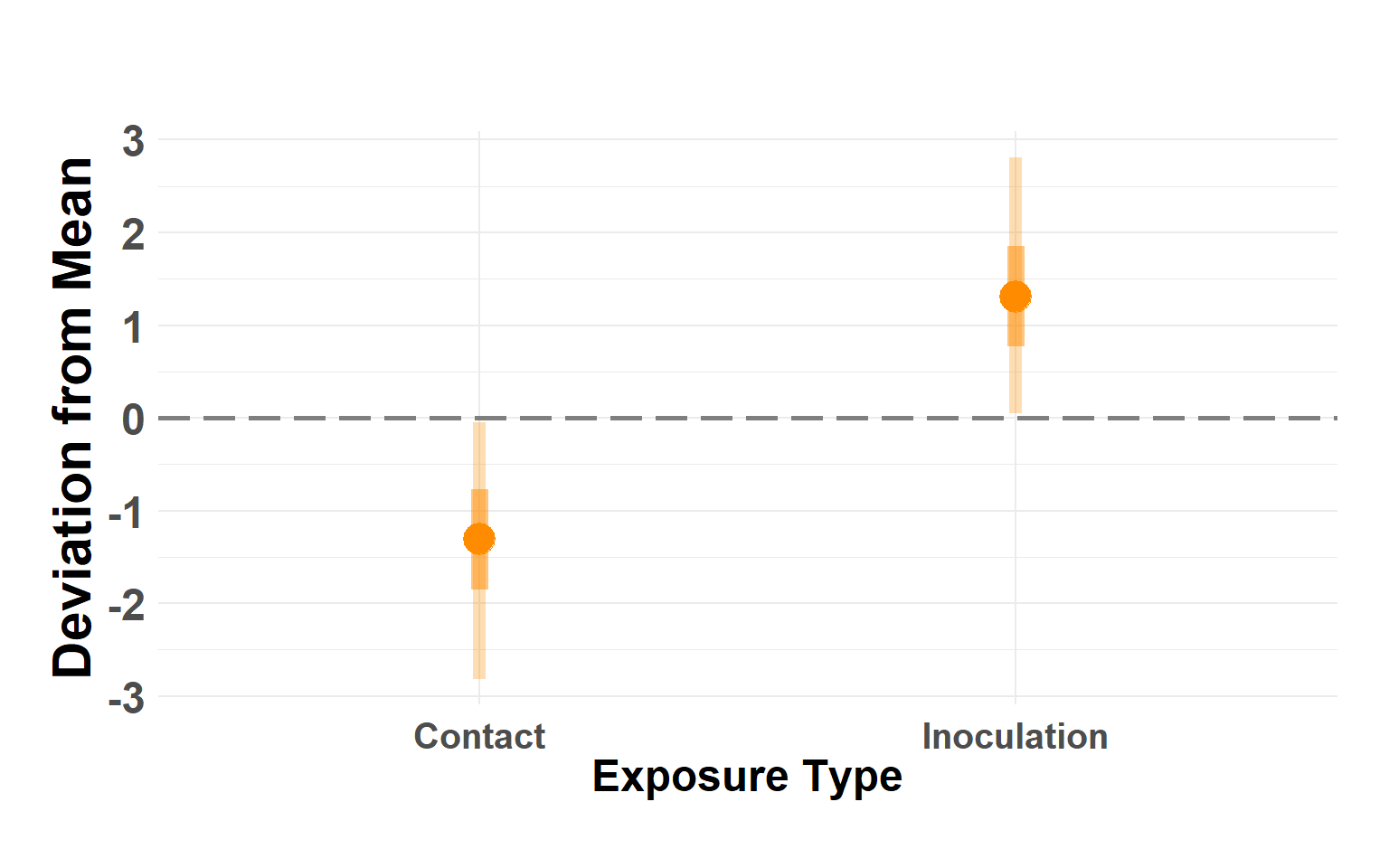
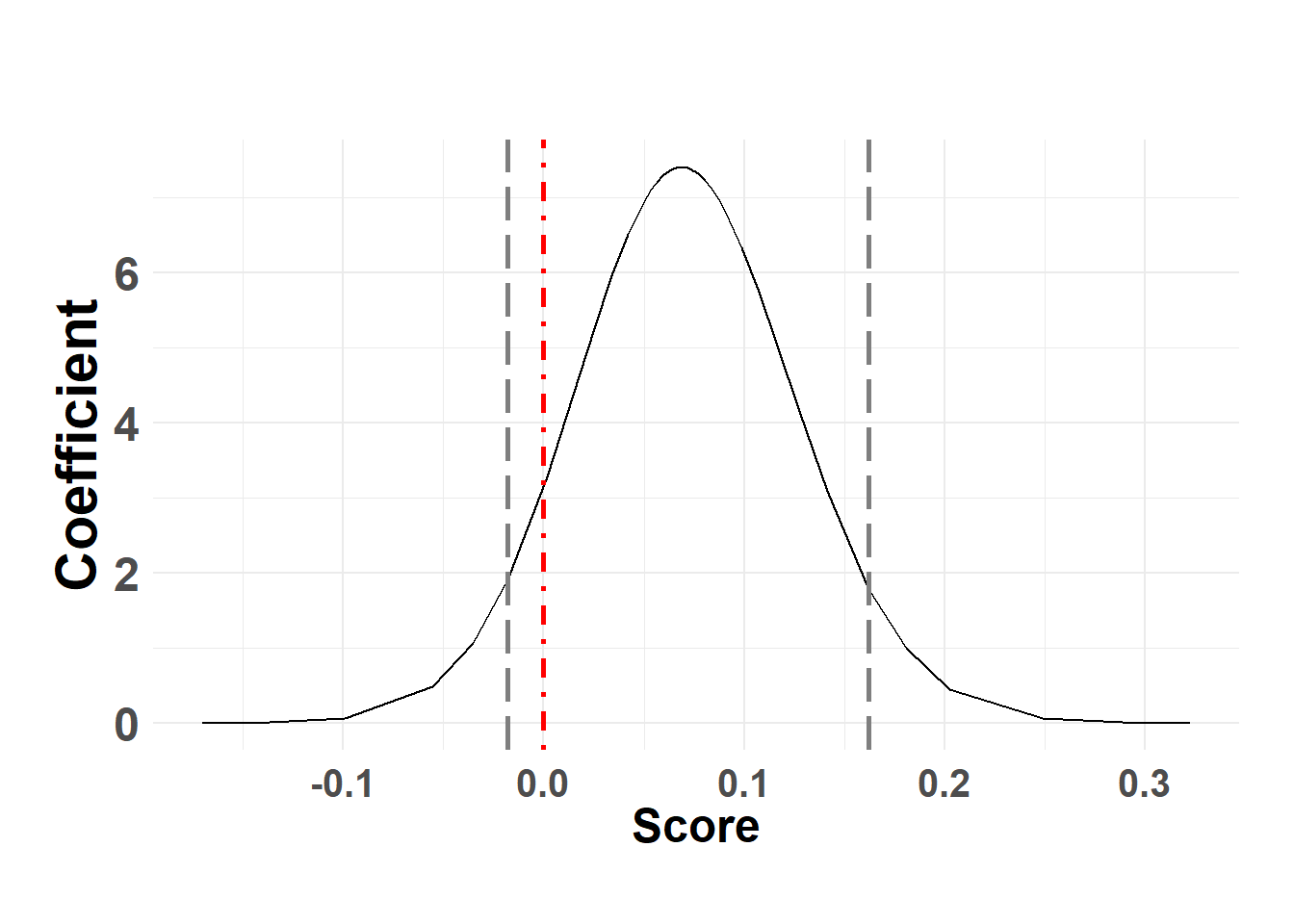
Fixed effects coefficients
Hide code
temp_post <- shed_model$marginals.fixed
names(temp_post)[1] "(Intercept)" "temp" Temperature
Hide code
plot_fixed_posterior(temp_post, model = shed_model, xlabel = "Temperature", fixed_eff = "temp")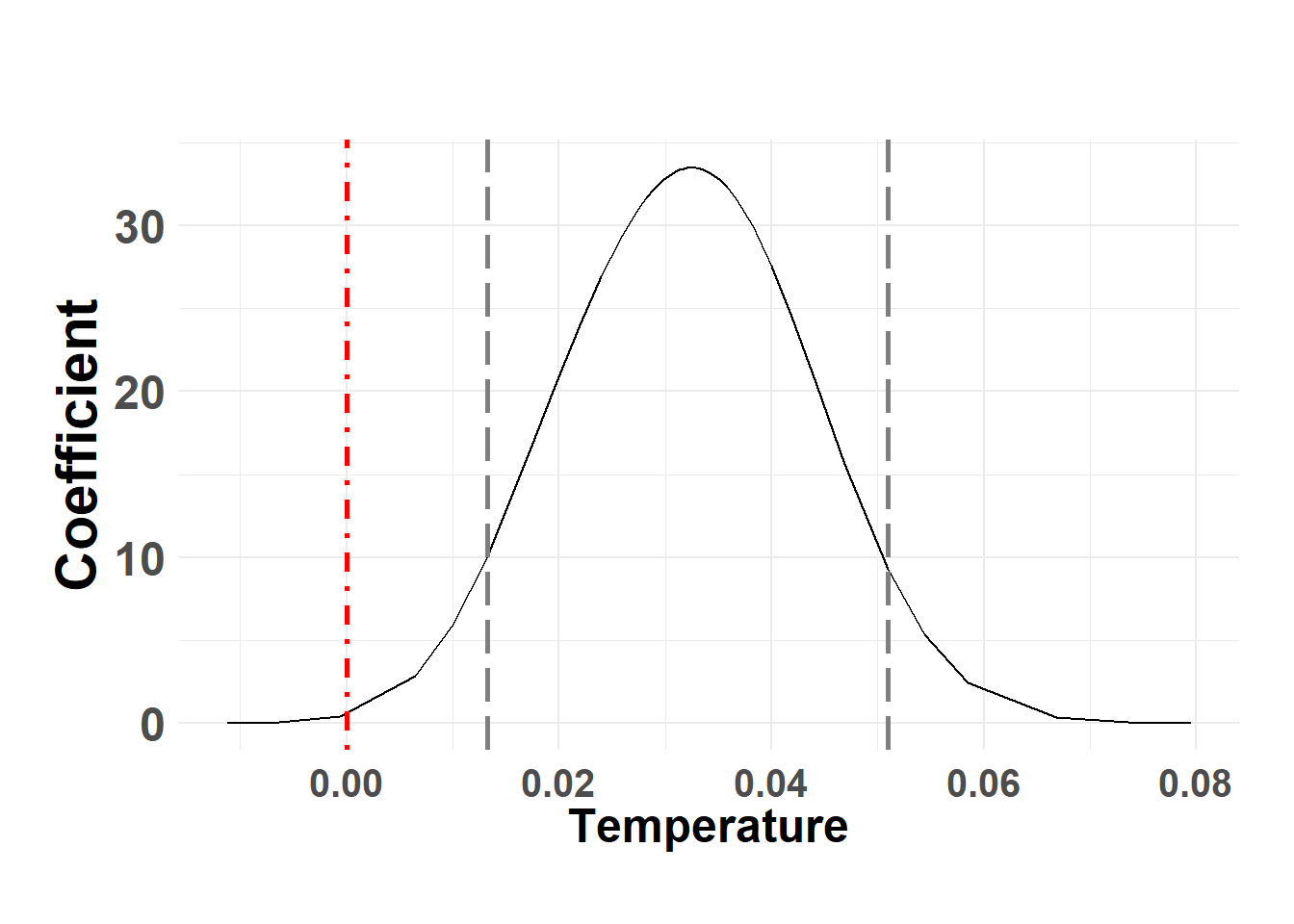
Intercept
Hide code
plot_fixed_posterior(temp_post, model = shed_model, xlabel = "Intercept", fixed_eff = "(Intercept)")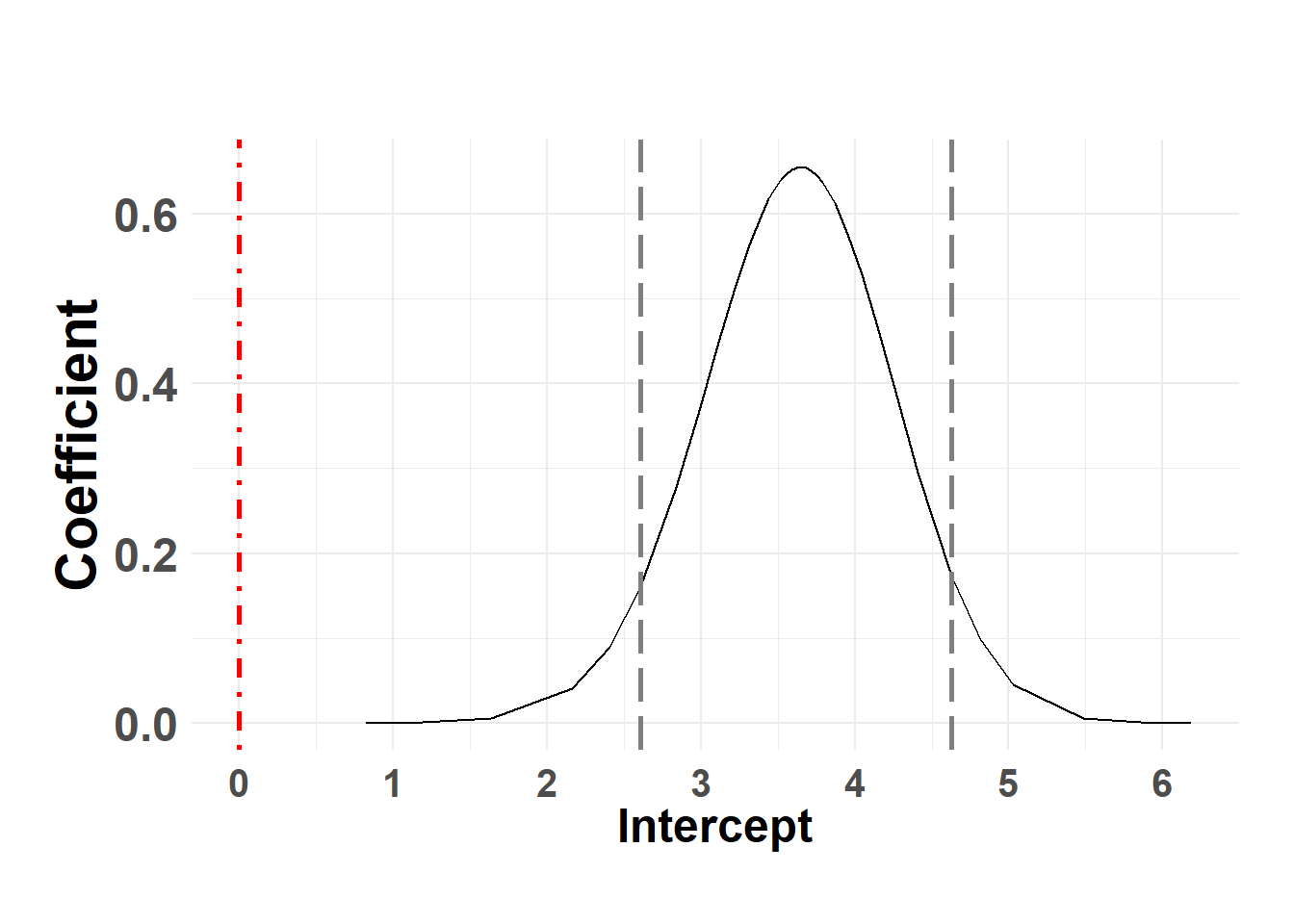
Serum
Run model for serum
Hide code
pc_prec_rw <- list(theta = list(prior="pc.prec",
param=c(1, 0.001)))
pc_prec_iid <- list(theta = list(prior="pc.prec",
param=c(3, 0.001)))
serum_form <- y_serum ~ 1 + f(time_step,
model = "rw1",
constr = TRUE,
scale.model = TRUE,
hyper=pc_prec_rw) +
f(animal,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
f(exp_type,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
temp_sc
serum_model <- inla(serum_form,
data = longitud_df,
family = "gaussian",
quantiles = return_quants,
verbose=FALSE,
control.compute=list(dic = TRUE, cpo = FALSE, waic = TRUE))
# summary(serum_model)Plot Time Effect
Hide code
time_df <- serum_model$summary.random$time_step[, c(1, 4:10)]
names(time_df) <- c("DPE", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
time_df$DPE <- time_df$DPE - 1
plot_time_rw(time_df)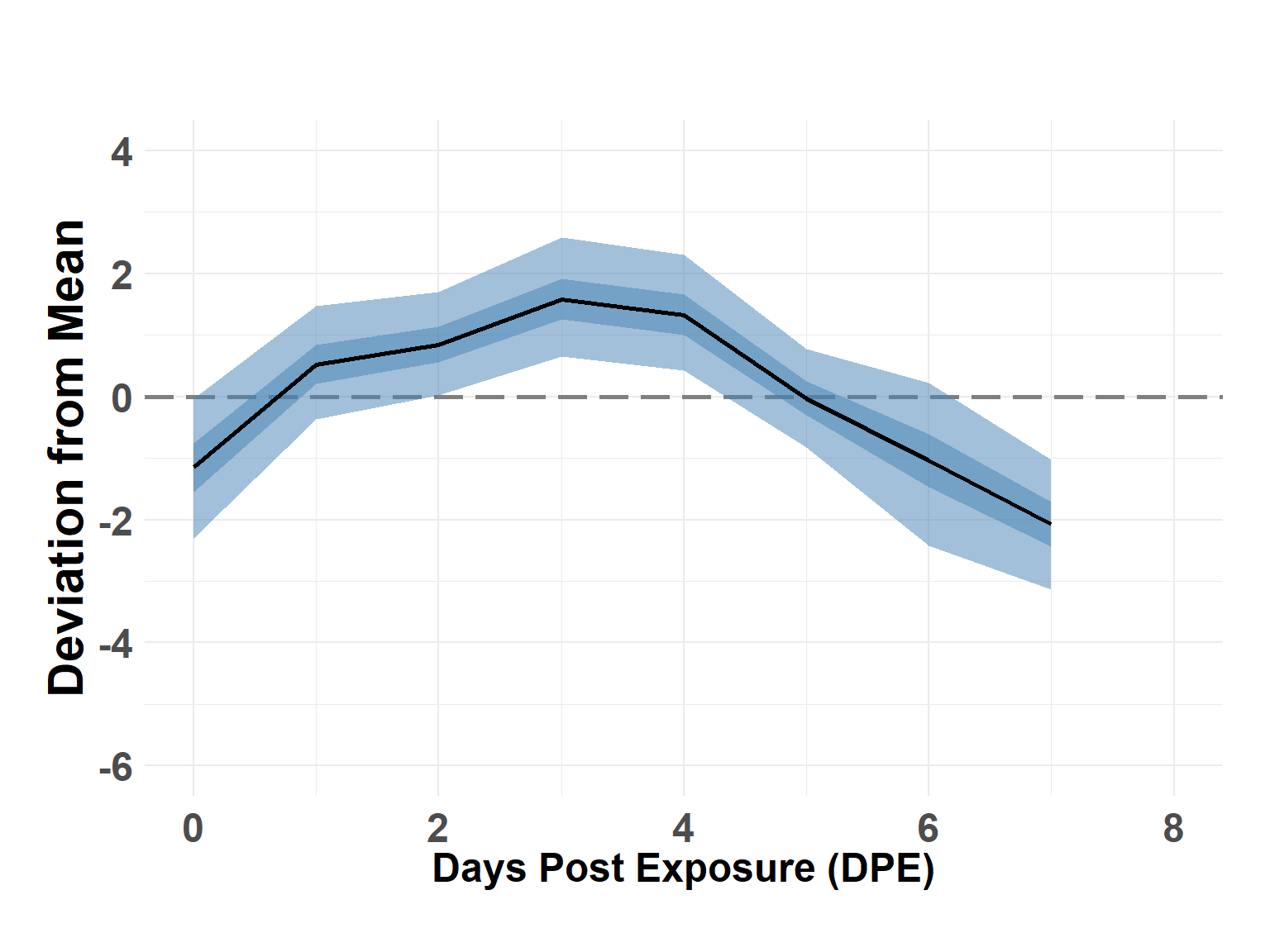
Plot animals
Hide code
animal_df <- serum_model$summary.random$animal[, c(1, 4:10)]
names(animal_df) <- c("Animal", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
plot_animals_linerange(animal_df)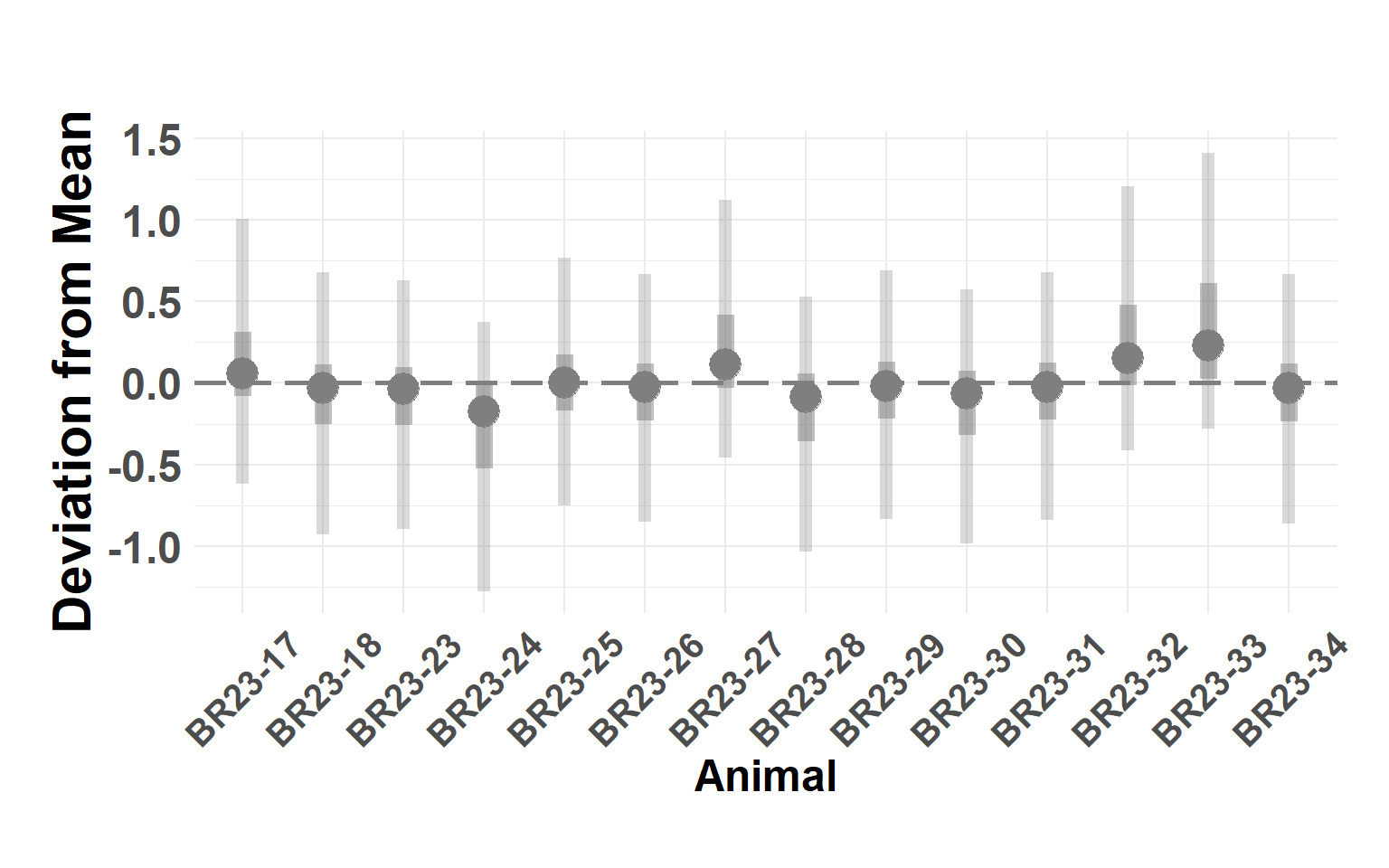
Plot exposure type
Hide code
exptype_df <- serum_model$summary.random$exp_type[, c(1, 4:10)]
names(exptype_df) <- c("type", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
exptype_df$type <- as.factor(exptype_df$type)
levels(exptype_df$type) <- c("Contact", "Inoculation")
plot_exposure_linerange(exptype_df)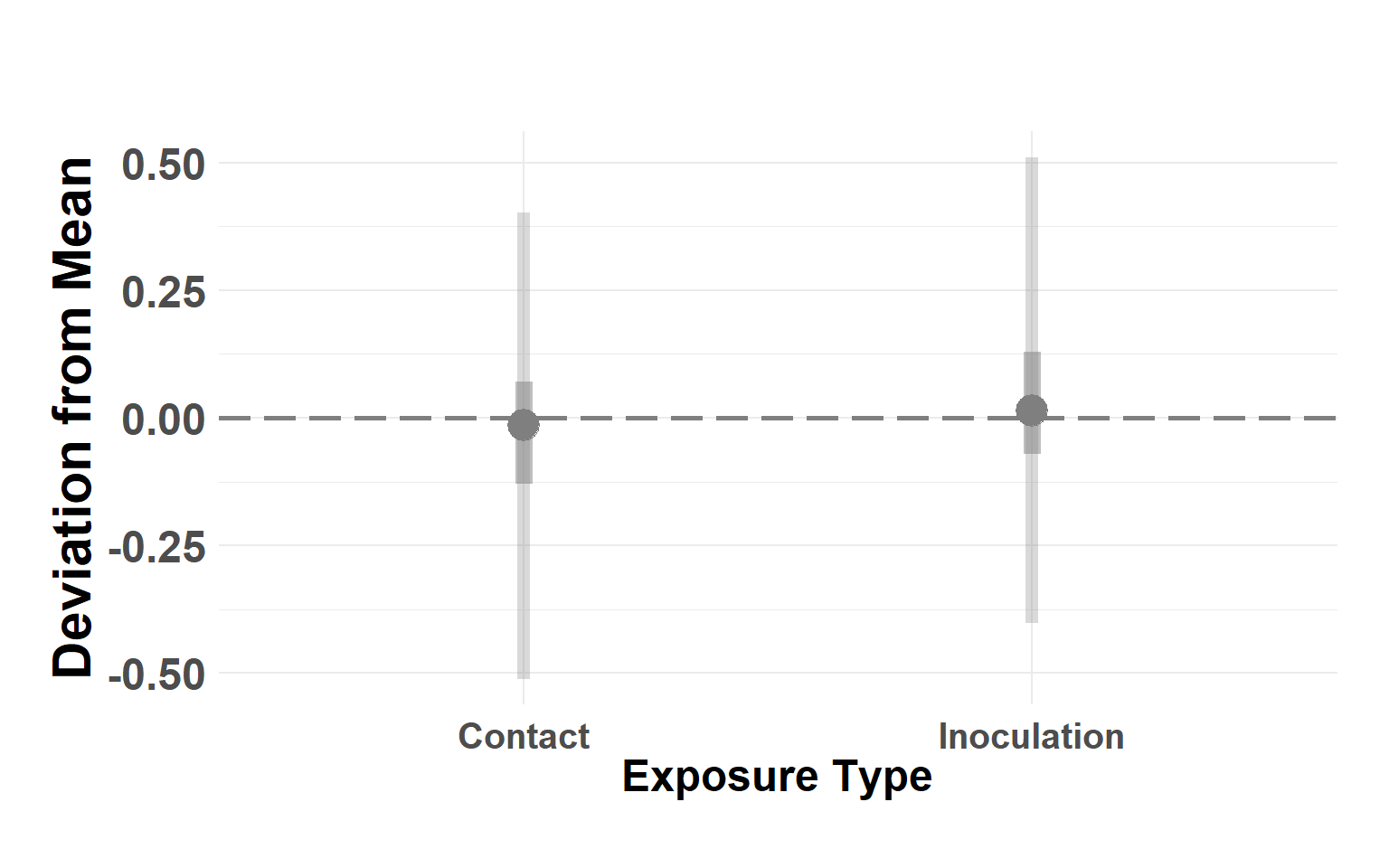
Fixed effects coefficients
Hide code
temp_post <- serum_model$marginals.fixed
names(temp_post)[1] "(Intercept)" "temp_sc" Temperature
Hide code
plot_fixed_posterior(temp_post, model = serum_model, xlabel = "Temperature", fixed_eff = "temp_sc")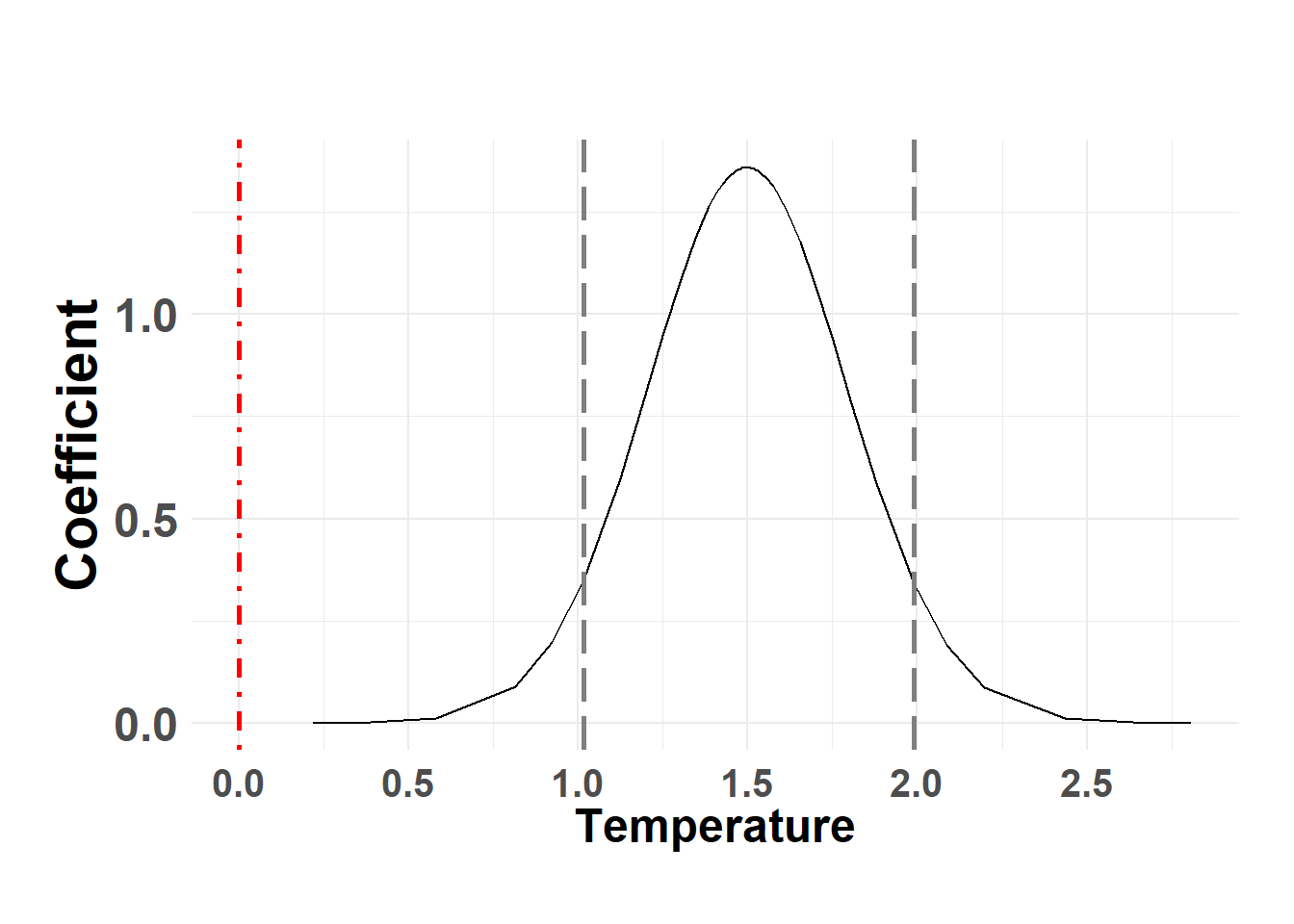
Intercept
Hide code
plot_fixed_posterior(temp_post, model = serum_model, xlabel = "Intercept", fixed_eff = "(Intercept)")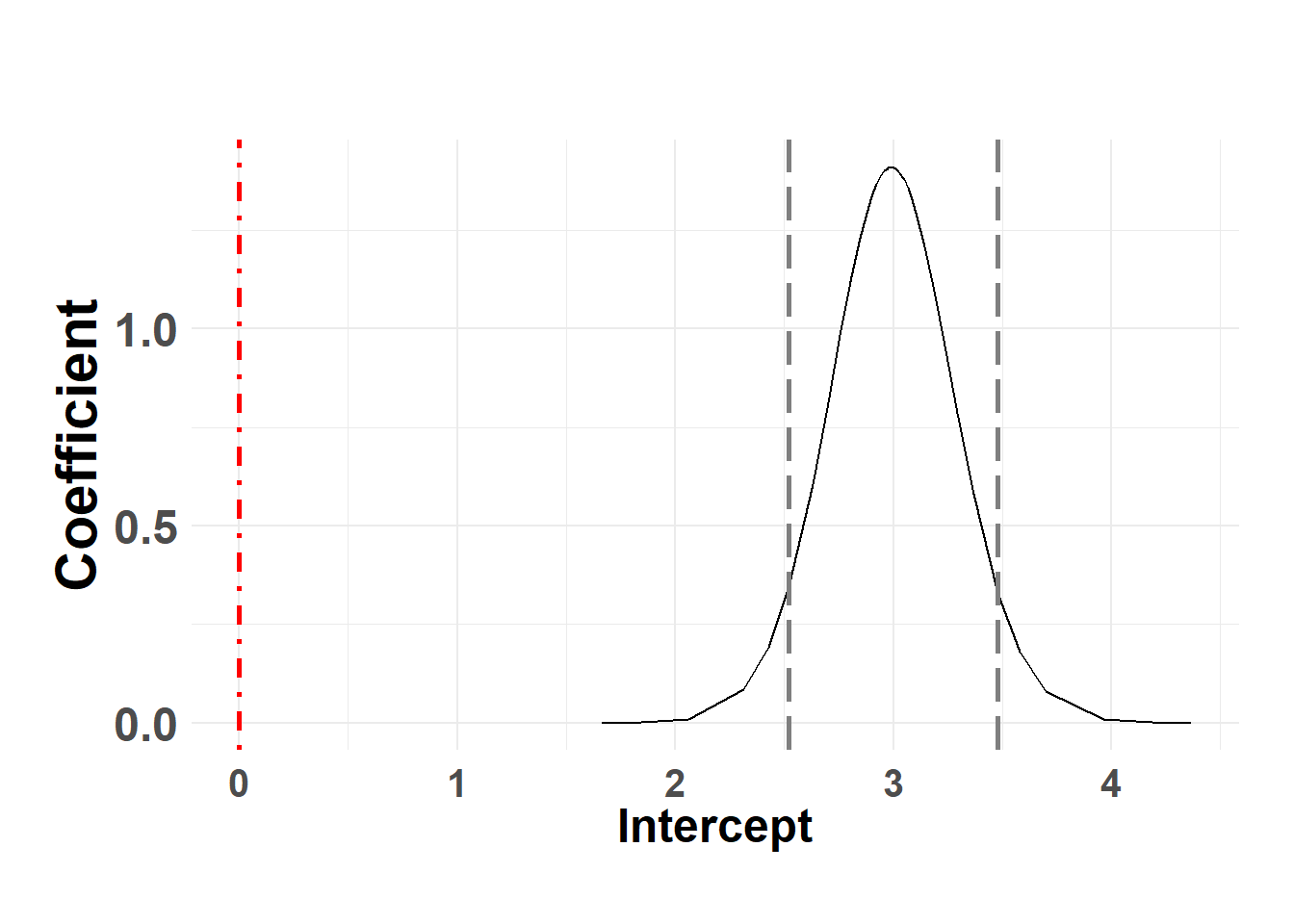
Lesion Score
Run model for lesion score
Hide code
pc_prec_rw <- list(theta = list(prior="pc.prec",
param=c(1, 0.001)))
pc_prec_iid <- list(theta = list(prior="pc.prec",
param=c(3, 0.001)))
score_form <- score ~ 1 + f(time_step,
model = "rw1",
constr = TRUE,
scale.model = TRUE,
hyper=pc_prec_rw) +
f(animal,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
f(exp_type,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
temp
score_model <- inla(score_form,
data = longitud_df,
family = "gaussian",
quantiles = return_quants,
verbose=FALSE,
control.compute=list(dic = TRUE, cpo = FALSE, waic = TRUE))
# summary(score_model)Plot Time Effect
Hide code
time_df <- score_model$summary.random$time_step[, c(1, 4:10)]
names(time_df) <- c("DPE", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
time_df$DPE <- time_df$DPE - 1
plot_time_rw(time_df)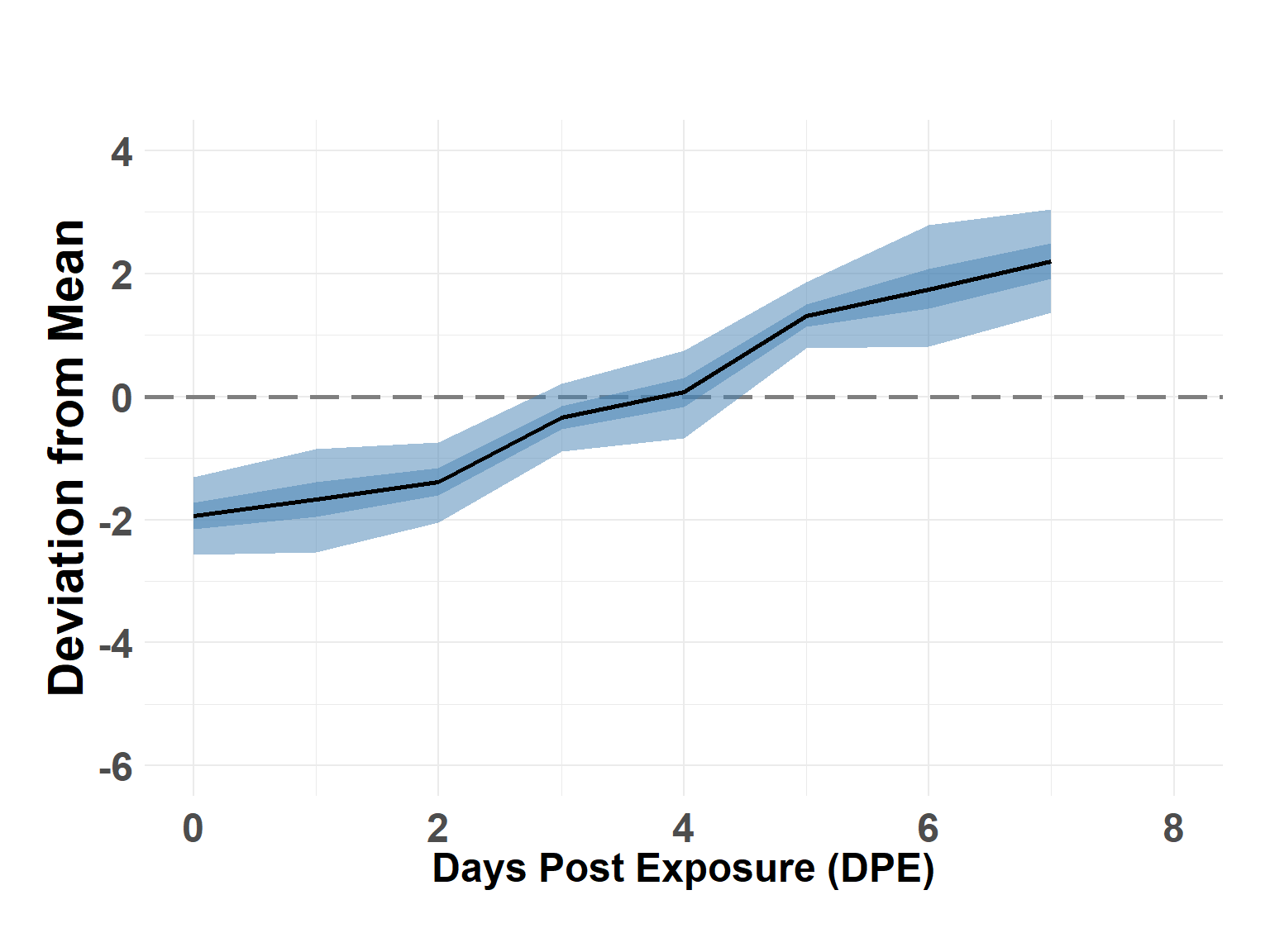
Plot animals
Hide code
animal_df <- score_model$summary.random$animal[, c(1, 4:10)]
names(animal_df) <- c("Animal", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
plot_animals_linerange(animal_df)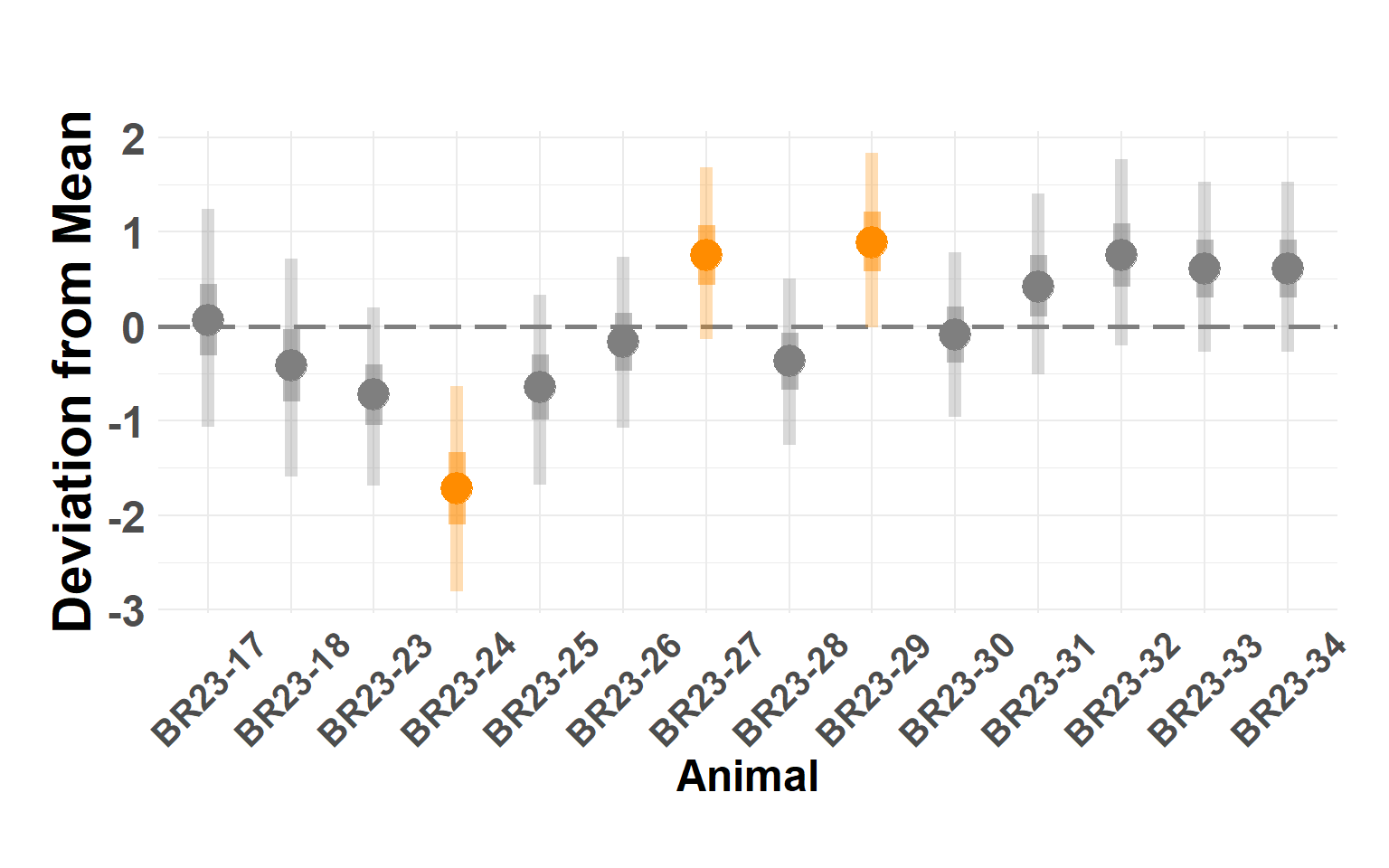
Plot exposure type
Hide code
exptype_df <- score_model$summary.random$exp_type[, c(1, 4:10)]
names(exptype_df) <- c("type", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
exptype_df$type <- as.factor(exptype_df$type)
levels(exptype_df$type) <- c("Contact", "Inoculation")
plot_exposure_linerange(exptype_df)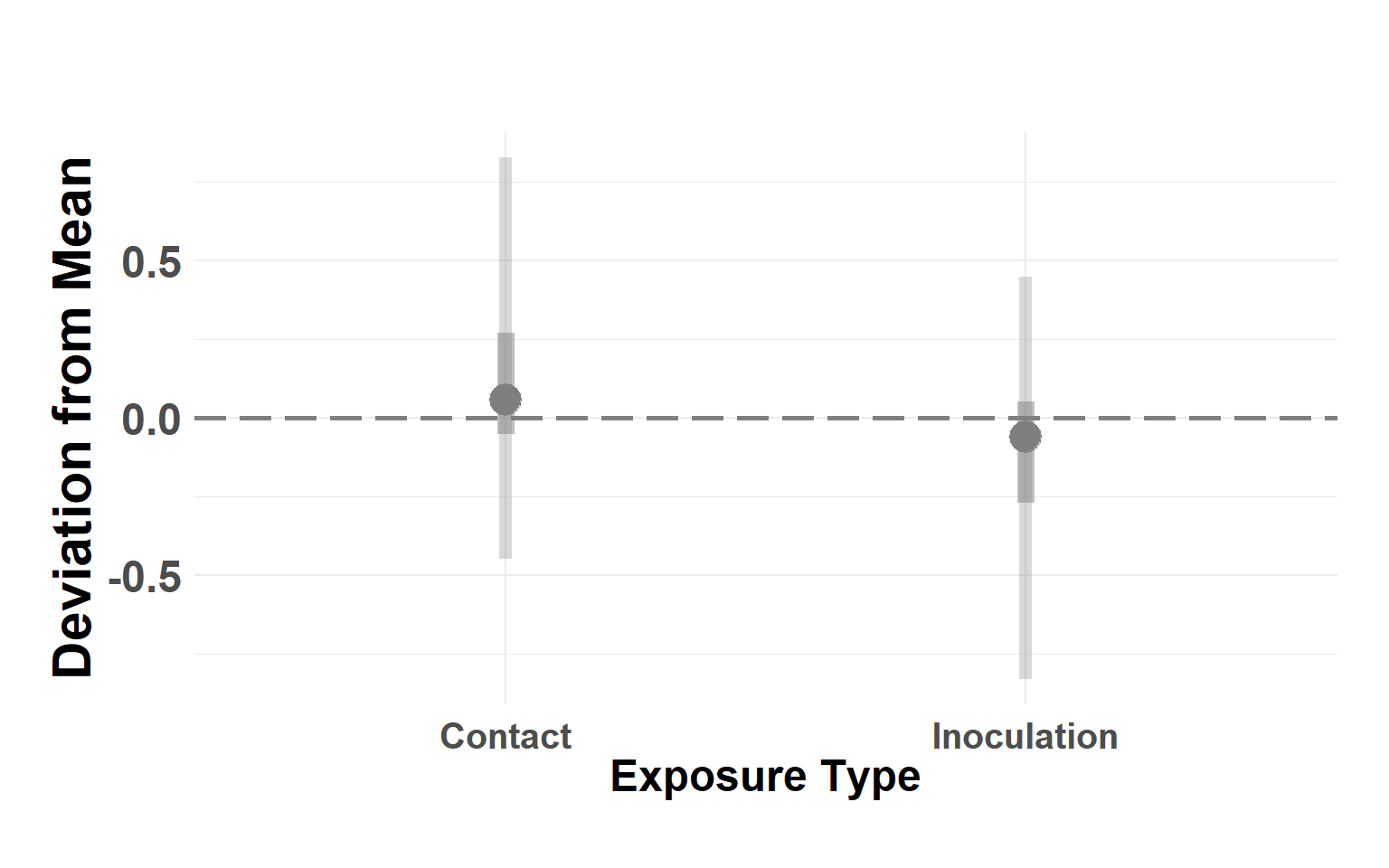
Fixed effects coefficients
Hide code
temp_post <- score_model$marginals.fixed
names(temp_post)[1] "(Intercept)" "temp" Temperature
Hide code
plot_fixed_posterior(temp_post, model = score_model, xlabel = "Temperature", fixed_eff = "temp")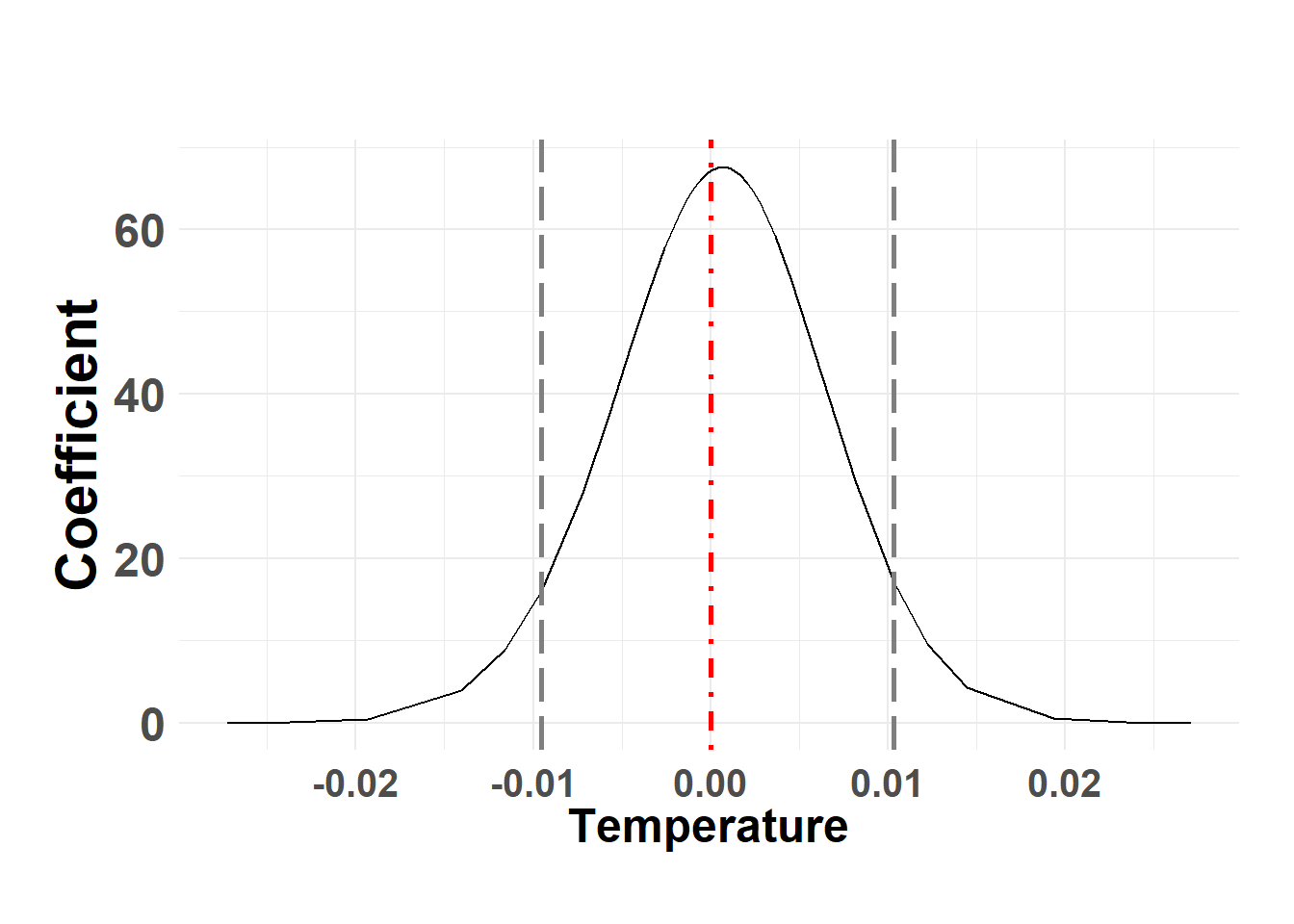
Intercept
Hide code
plot_fixed_posterior(temp_post, model = score_model, xlabel = "Intercept", fixed_eff = "(Intercept)")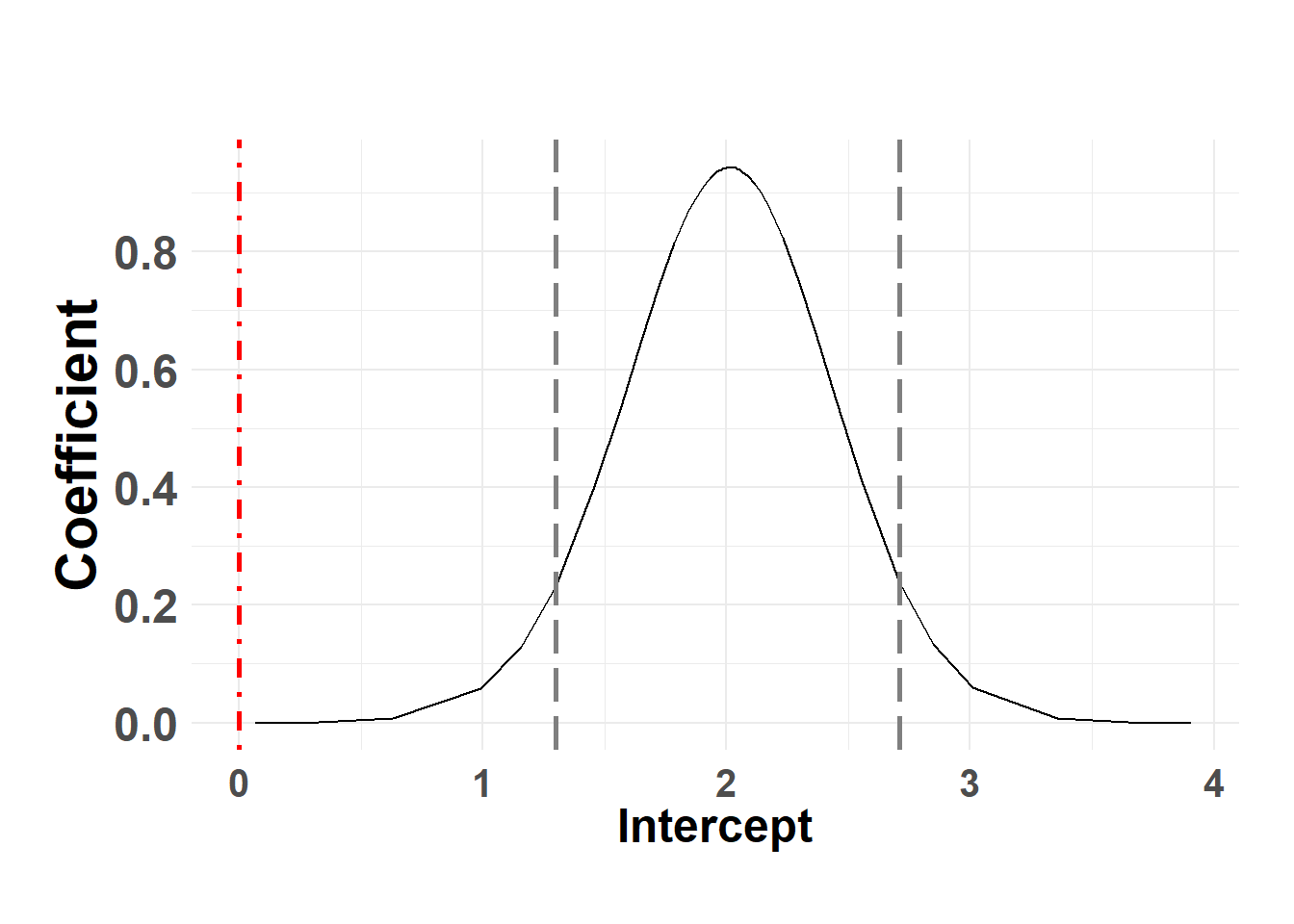
Temperature
Run model for body temperature
Hide code
longitud_df$y_temper <- as.numeric(scale(longitud_df$temp, scale=TRUE, center=TRUE))
pc_prec_rw <- list(theta = list(prior="pc.prec",
param=c(1, 0.001)))
pc_prec_iid <- list(theta = list(prior="pc.prec",
param=c(3, 0.001)))
temper_form <- y_temper ~ -1 + f(time_step,
model = "rw1",
constr = TRUE,
scale.model = TRUE,
hyper=pc_prec_rw) +
f(animal,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
f(exp_type,
model = "iid",
constr=TRUE,
hyper=pc_prec_iid) +
score
temper_model <- inla(temper_form,
data = longitud_df,
family = "gaussian",
quantiles = return_quants,
verbose=FALSE,
control.compute=list(dic = TRUE, cpo = FALSE, waic = TRUE))
# summary(temper_model)Plot Time Effect
Hide code
time_df <- temper_model$summary.random$time_step[, c(1, 4:10)]
names(time_df) <- c("DPE", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
time_df$DPE <- time_df$DPE - 1
plot_time_rw(time_df)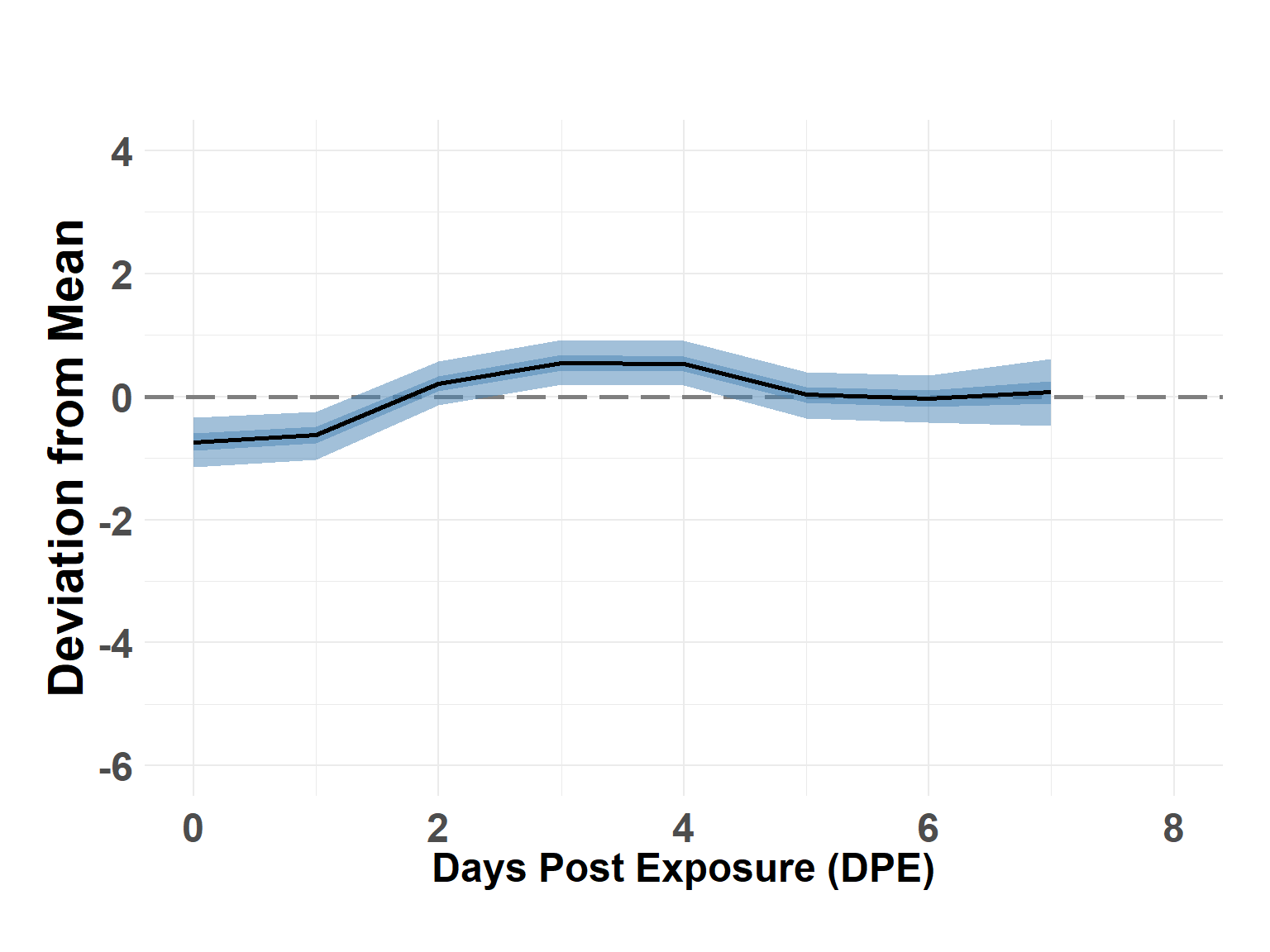
Plot animals
Hide code
animal_df <- temper_model$summary.random$animal[, c(1, 4:10)]
names(animal_df) <- c("Animal", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
plot_animals_linerange(animal_df)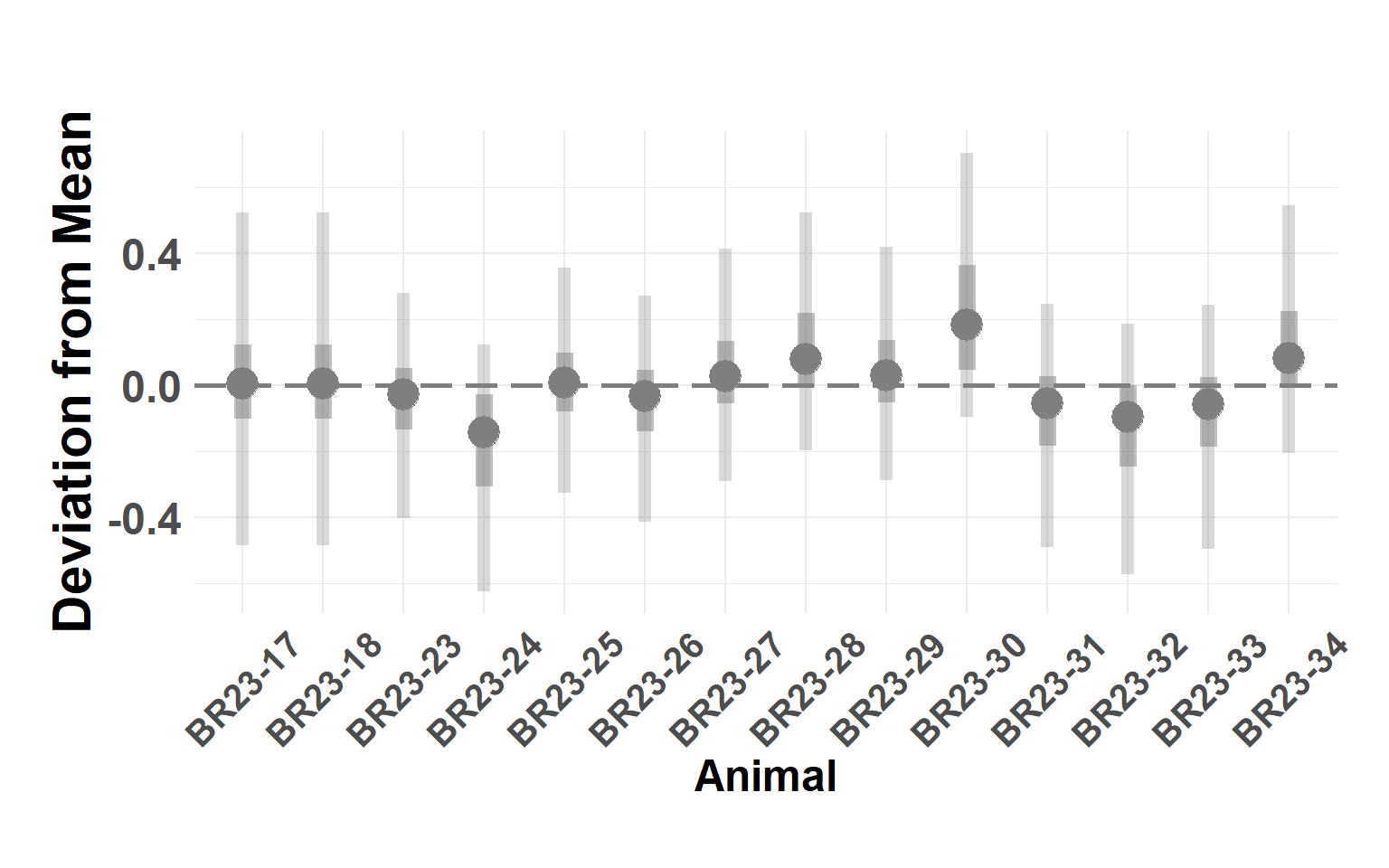
Plot exposure type
Hide code
exptype_df <- temper_model$summary.random$exp_type[, c(1, 4:10)]
names(exptype_df) <- c("type", "Q_0.025", "Q_0.05", "Q_0.25", "Q_0.5", "Q_0.75", "Q_0.95", "Q_0.975")
exptype_df$type <- as.factor(exptype_df$type)
levels(exptype_df$type) <- c("Contact", "Inoculation")
plot_exposure_linerange(exptype_df)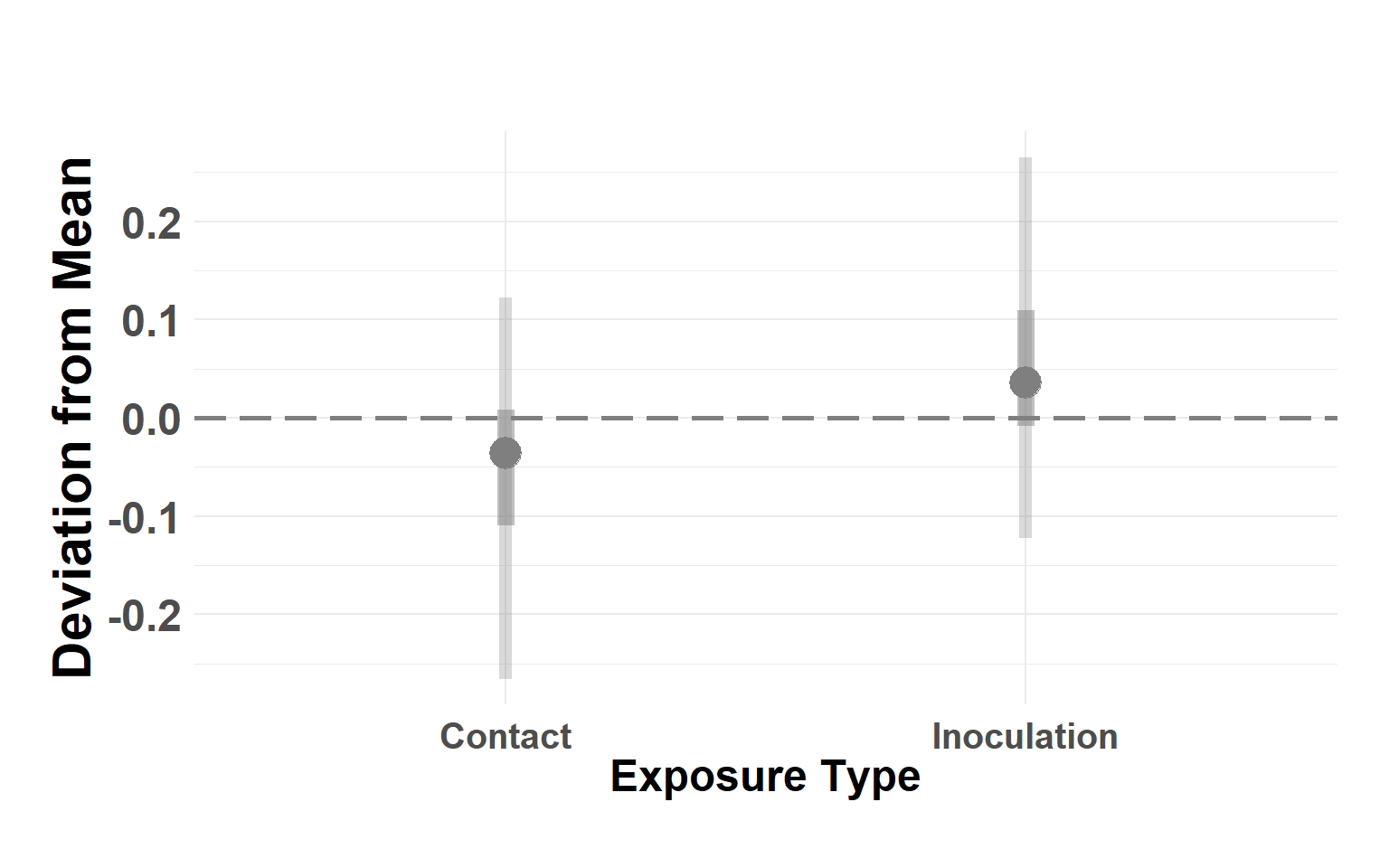
Fixed effects coefficients
Hide code
temp_post <- temper_model$marginals.fixed
names(temp_post)[1] "score"Score
Hide code
plot_fixed_posterior(temp_post, model = temper_model, xlabel = "Score", fixed_eff = "score")